Advancing Food Safety: Electrochemical Biosensor Protocols for Rapid Detection of Pesticide Residues in Fruits
This article provides a comprehensive overview of electrochemical biosensor technology for the detection of pesticide residues in fruits, tailored for researchers and scientists in food safety and drug development.
Advancing Food Safety: Electrochemical Biosensor Protocols for Rapid Detection of Pesticide Residues in Fruits
Abstract
This article provides a comprehensive overview of electrochemical biosensor technology for the detection of pesticide residues in fruits, tailored for researchers and scientists in food safety and drug development. It covers the foundational principles of pesticide toxicity and biosensor operation, delves into detailed methodological protocols for sensor fabrication and application, addresses critical troubleshooting and optimization strategies for real-world samples, and presents a rigorous validation framework comparing biosensor performance against traditional chromatographic methods. The content synthesizes the latest advancements from 2024-2025, highlighting the transition toward portable, on-site analysis for enhanced food safety monitoring within a 'One Health' context.
The Urgent Need for Rapid Pesticide Detection: Principles and Public Health Imperatives
The widespread application of organophosphates (OPs), carbamates, and organochlorines in agriculture has made their residue detection in food products a critical public health concern. These compounds are designed to be biologically active and can inhibit essential nervous system enzymes in pests, but they pose significant risks to human health through the consumption of contaminated fruits and vegetables. Electrochemical biosensors have emerged as powerful tools for the rapid, sensitive, and cost-effective detection of these pesticide residues, offering a viable alternative to traditional chromatographic methods. This document provides detailed application notes and protocols for researchers developing these biosensing platforms, with a specific focus on assays for fruit samples.
Pesticide Classes: Mechanisms and Analytical Targets
The three pesticide classes of concern share a common neurotoxic mechanism but differ in their chemical structures, persistence, and specific modes of action. Understanding these differences is fundamental to designing specific detection protocols.
Table 1: Characteristics of Key Pesticide Classes
| Pesticide Class | Primary Mechanism of Action | Example Compounds | Key Structural/Functional Groups for Detection | Environmental Persistence |
|---|---|---|---|---|
| Organophosphates (OPs) | Irreversible inhibition of acetylcholinesterase (AChE) [1] [2] | Chlorpyrifos, Parathion, Dichlorvos, Malathion [3] [2] | P=O (Oxon) or P=S (Thion) groups; P-O-C bonds [2] [4] | Low to moderate [2] |
| Carbamates | Reversible inhibition of acetylcholinesterase (AChE) [1] [2] | Carbaryl, Carbofuran, Methomyl, Aldicarb [1] [2] | Carbamate ester group (OC(O)N) [2] | Low [2] |
| Organochlorines | Disruption of sodium and potassium channels in neurons [5] | DDT, Endosulfan, Lindane [5] | Chlorinated cyclic hydrocarbons [5] | High (Persistent Organic Pollutants) [5] |
The following diagram illustrates the shared signaling pathway through which organophosphates and carbamates exert their neurotoxic effect, which is the basis for many enzyme-based biosensors.
A variety of biosensing platforms have been developed for pesticide detection, each with distinct performance characteristics. The table below summarizes the analytical performance of different transducer types as reported in recent literature.
Table 2: Comparison of Biosensor Platforms for Pesticide Detection
| Transducer Type | Target Pesticide (Example) | Reported Limit of Detection (LOD) | Linear Range | Key Advantages |
|---|---|---|---|---|
| Electrochemical (AChE-based) | Carbofuran [6] | Not specified in excerpt | Not specified in excerpt | High sensitivity, cost-effective, portable [7] [6] |
| Piezoelectric (QCM) | Carbaryl [2] | 2 × 10⁻¹⁰ M [2] | Not specified in excerpt | Label-free, real-time output, high sensitivity [2] |
| Piezoelectric (QCM) | Diisopropylfluorophosphate [2] | 1 × 10⁻¹⁰ M [2] | Not specified in excerpt | Label-free, real-time output, high sensitivity [2] |
| Electrochemical (MIP-based) | Captan [8] | 1 × 10⁻¹⁴ M [8] | 1 × 10⁻¹⁴ to 9 × 10⁻¹⁴ M [8] | High selectivity, enzyme-free stability, reusability [8] |
| Cell-based (Bioelectric) | Chlorpyrifos & Carbaryl [1] | 1 ppb (approx. 10⁻⁹ M) [1] | Not specified in excerpt | Measures functional physiological response (cell membrane potential) [1] |
Detailed Experimental Protocols
This section provides a step-by-step workflow for two primary electrochemical biosensor protocols: one based on enzyme inhibition and another utilizing molecularly imprinted polymers.
Protocol 1: AChE-based Electrochemical Biosensor for Fruit Samples
This protocol is adapted from established AChE-sensor methodologies for detecting organophosphate and carbamate pesticides [1] [6].
4.1.1 Workflow Diagram
4.1.2 Materials and Reagents
- Acetylcholinesterase (AChE): Enzyme biorecognition element, typically from electric eel [6].
- Acetylthiocholine (ATCh) or Acetylcholine (ACh): Enzyme substrate; hydrolysis produces electroactive product [1] [6].
- Working Electrode: Often Glassy Carbon Electrode (GCE), potentially modified with nanomaterials (e.g., carbon black) to enhance sensitivity [6].
- Chitosan/Nafion: Polymers used for enzyme immobilization and membrane formation on the electrode surface [6].
- Electrochemical Cell: Standard three-electrode setup (Working, Reference Ag/AgCl, Auxiliary Pt electrode) [8].
- Buffer Solutions: Phosphate or Tris buffer for maintaining optimal pH (e.g., pH 8 for AChE activity) [1].
- Extraction Solvent: Aqueous-organic mixture (e.g., water:acetone 1:3 v/v) for pesticide extraction from fruit [1].
4.1.3 Step-by-Step Procedure
- Sample Preparation: Homogenize fruit sample (e.g., apple, tomato). Extract pesticides using a suitable solvent like water:acetone (1:3 v/v). Evaporate acetone and use the aqueous supernatant for analysis [1] [8].
- Biosensor Fabrication: Immobilize AChE onto the surface of the working electrode. This can be done by drop-casting a mixture of the enzyme and a binder like chitosan or Nafion, followed by cross-linking with glutaraldehyde to stabilize the enzyme layer [6].
- Baseline Enzyme Activity Measurement: Immerse the biosensor in a buffer containing the substrate (e.g., acetylthiocholine). Measure the amperometric or voltammetric (DPV) current generated by the enzymatic product (thiocholine). This signal represents 100% enzyme activity [6].
- Inhibition/Incubation Step: Incubate the biosensor with the prepared fruit extract for a fixed time (e.g., 10-30 minutes). Any OPs or carbamates present will inhibit the AChE enzyme [1] [6].
- Post-Inhibition Activity Measurement: Re-immerse the sensor in the substrate solution and measure the electrochemical signal again. The signal will be lower due to enzyme inhibition [6].
- Quantification: Calculate the percentage of enzyme inhibition:
% Inhibition = [(I₀ - I₁) / I₀] × 100, where I₀ is the baseline current and I₁ is the current after exposure. The pesticide concentration is determined by comparing the % inhibition to a calibration curve prepared with known pesticide standards [6].
Protocol 2: Molecularly Imprinted Polymer (MIP)-based Electrochemical Sensor
This protocol details a non-enzymatic approach for specific pesticide detection, using Captan as an example [8].
4.2.1 Workflow Diagram
4.2.2 Materials and Reagents
- Template Molecule: The target pesticide (e.g., Captan) [8].
- Functional Monomer:
o-Phenylenediamine (o-PD) [8]. - Cross-linker & Electrolyte: Acetate buffer (pH 5.2) [8].
- Electrochemical Cell: Three-electrode system with GCE as working electrode [8].
- Extraction Solution: 1M HCl and Acetonitrile (ACN) mixture (7:3 v/v) [8].
- Redox Probe: Potassium ferrocyanide/ferricyanide ([Fe(CN)₆]³⁻/⁴⁻) in KCl [8].
4.2.3 Step-by-Step Procedure
- MIP Fabrication (Electropolymerization): Immerse a clean GCE in a solution containing the functional monomer (
o-PD), the template (Captan), and acetate buffer. Perform Cyclic Voltammetry (CV) for a set number of cycles (e.g., 10 cycles between -0.2 V and 0.8 V) to electropolymerize theo-PD around the template molecules [8]. - Template Extraction: Wash the polymer-coated electrode (MIP@o-PD/GCE) with a mixture of 1M HCl and acetonitrile to remove the Captan template molecules. This leaves specific recognition cavities in the polymer matrix [8].
- Analyte Rebinding: Incubate the MIP sensor in the prepared fruit extract. Captan molecules from the sample will selectively rebind to the cavities, changing the properties of the polymer layer [8].
- Electrochemical Measurement: Measure the Differential Pulse Voltammetry (DPV) response of the sensor in a solution containing the [Fe(CN)₆]³⁻/⁴⁻ redox probe. The rebinding of Captan impedes the access of the redox probe to the electrode surface, causing a decrease in the peak current [8].
- Quantification: The decrease in DPV peak current is proportional to the concentration of Captan in the sample. A calibration curve is constructed using standards to enable quantification [8].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagents and Materials for Electrochemical Biosensor Development
| Item | Function/Application | Example from Literature |
|---|---|---|
| Acetylcholinesterase (AChE) | Primary biorecognition element for OP and carbamate detection via enzyme inhibition [1] [6]. | Electric eel AChE [6] |
| Acetylthiocholine (ATCh) / Acetylcholine (ACh) | Enzyme substrate; hydrolysis produces an electroactive product (thiocholine/choline) for signal generation [1] [6]. | Acetylthiocholine iodide (ATChI) [6] |
| Glassy Carbon Electrode (GCE) | Common working electrode substrate; provides a stable, conductive surface for enzyme/MIP immobilization [8]. | 3 mm diameter GCE [8] |
| Molecularly Imprinted Polymer (MIP) | Synthetic polymer with specific cavities for target analytes; offers enzyme-free, stable recognition [8]. | o-Phenylenediamine (o-PD) polymer for Captan [8] |
| Nanomaterial Modifiers (e.g., Carbon Black) | Enhance electrode surface area, improve electron transfer, and increase biosensor sensitivity [6]. | Conductive carbon black (Vulcan XC 72R) [6] |
| Immobilization Matrices (Chitosan, Nafion) | Form stable membranes on electrodes to entrap biorecognition elements (enzymes) [6]. | Chitosan & Nafion used for AChE immobilization [6] |
| Redox Probes (e.g., [Fe(CN)₆]³⁻/⁴⁻) | Used with impedimetric or voltammetric techniques to monitor binding events or insulating layer formation on the electrode surface [8]. | Potassium ferrocyanide/ferricyanide [8] |
Critical Considerations for Assay Development
- Matrix Effects: The complex composition of fruit extracts can significantly interfere with sensor performance. Components like lipids, organic acids, or sugars can foul the electrode or non-specifically inhibit enzymes. Calibration curves should be constructed in the presence of a blank (pesticide-free) extract of the same fruit matrix to account for these effects [6].
- Sensor Regeneration: For reusable sensors, a regeneration step is required to remove the bound pesticide from the biorecognition element. For AChE-based sensors, this can be challenging due to the irreversible binding of OPs. For MIP sensors, the template extraction protocol can often be used for regeneration between measurements [8] [6].
- Multiplexed Detection: A significant challenge is detecting specific pesticides within a class or differentiating between OPs and carbamates. Using arrays of sensors with different biorecognition elements (e.g., AChE from different species, specific antibodies, or multiple MIPs) coupled with advanced data analysis is a promising approach to address this challenge [4].
Pesticide residues on agricultural products present a significant global public health challenge due to their potential to cause both immediate poisoning and long-term neurological damage. The widespread application of pesticides in modern agriculture has made residue exposure an unavoidable concern for consumers worldwide [9]. Understanding the dual-toxicity profile—acute effects following high-dose exposure and chronic neurodegenerative consequences from prolonged low-level exposure—is paramount for developing effective safety monitoring protocols. Electrochemical biosensors represent a transformative technology for quantifying these risks, offering rapid, sensitive, and field-deployable analysis of pesticide residues directly on food surfaces [10] [11]. This document provides detailed application notes and experimental protocols for assessing health risks from pesticide residues, with specific methodologies adapted for integration into electrochemical biosensing platforms for fruit analysis.
Health Risk Assessment of Pesticide Residues
Acute Toxicity Profiles of Major Pesticide Classes
Table 1: Acute Toxicity Mechanisms and Health Effects of Major Pesticide Classes
| Pesticide Class | Representative Compounds | Primary Mechanism of Action | Acute Health Effects | Detection Priority |
|---|---|---|---|---|
| Organophosphorus | Parathion, Dichlorvos, Malathion | Irreversible inhibition of acetylcholinesterase (AChE) [12] | Dyspnea, pulmonary edema, muscle spasms, dizziness, headache, bradycardia [12] | High (Enzyme Inhibition) |
| Carbamates | Carbaryl, Aldicarb, Carbofuran | Reversible inhibition of acetylcholinesterase (AChE) [12] | Dizziness, blurred vision, nausea, vomiting, abdominal pain, excessive sweating, tremors [12] | High (Enzyme Inhibition) |
| Neonicotinoids | Imidacloprid, Thiamethoxam | Continuous activation of nicotinic acetylcholine receptors [12] | Nausea, vomiting, headache, dizziness, insomnia, anxiety, consciousness disorders [12] | Medium (Receptor Binding) |
| Pyrethroids | Permethrin, Deltamethrin, Fenvalerate | Interference with sodium channels and GABA receptors [12] | Skin rashes, nausea, abdominal pain, headache, confusion, cough [12] | Medium (Biomimetic Assay) |
| Organochlorines | Hexachlorocyclohexane, Toxaphene | Inhibition of GABA receptors, ROS generation, endocrine disruption [12] | Similar to pyrethroids, plus endocrine disorders [12] | Low (Mostly Banned) |
Chronic Neurodegenerative Consequences
Beyond immediate toxicity, chronic exposure to certain pesticides, even at low concentrations, poses significant risks for neurodegenerative pathologies. The mechanisms involve complex interactions between genetic susceptibility and environmental exposures, where pesticides can act as neurotoxic stressors that accelerate or trigger pathological processes [13].
Key pathophysiological pathways include:
- Oxidative Stress: Many pesticides, including organochlorines, induce reactive oxygen species (ROS) generation, leading to neuronal oxidative damage and mitochondrial dysfunction [13] [12].
- Neuroinflammation: Chronic microglial activation by pesticide exposure creates a neuroinflammatory environment conducive to neurodegeneration [13].
- Protein Misfolding and Aggregation: Some pesticides can promote the misfolding and aggregation of proteins like α-synuclein and β-amyloid, hallmarks of Parkinson's and Alzheimer's diseases respectively [13].
- Epigenetic Modifications: Chronic exposure can alter DNA methylation and histone modification patterns, potentially leading to sustained changes in gene expression relevant to neuronal survival and function [13].
Epidemiological studies have consistently demonstrated associations between pesticide exposure and increased incidence of Parkinson's disease, Alzheimer's disease, and Amyotrophic Lateral Sclerosis (ALS) [13]. The delayed onset and progressive nature of these conditions make early detection of exposure biomarkers particularly critical for preventive interventions.
Experimental Protocols for Risk Assessment Using Biosensors
Protocol 1: On-Glove Electrochemical Detection of Organophosphorus Pesticides on Fruit Peels
Principle: This protocol describes a direct, on-site method for detecting organophosphorus pesticides (e.g., dichlorvos) on fruit surfaces using an enzymatic inhibition biosensor integrated onto a glove fingertip [10].
Workflow Overview:
Materials and Reagents:
- Butyrylcholinesterase Enzyme: Biological recognition element whose inhibition is proportional to pesticide concentration [10].
- Prussian Blue and Carbon Black Nanomaterials: Electron-transfer mediators for enhanced electrochemical signal transduction [10].
- Screen-Printed Electrodes (SPEs): Miniaturized, disposable sensing platforms integrated onto glove fingertips [10].
- Acetylthiocholine or Butyrylthiocholine: Enzyme substrates that produce electroactive products upon hydrolysis [10].
- Portable Potentiostat: Miniaturized electrochemical analyzer for field-deployable measurements [10].
- Protective Nitrile Gloves: Base platform for sensor integration.
Procedure:
- Biosensor Preparation: Fabricate screen-printed electrodes modified with Prussian blue, carbon black, and butyrylcholinesterase enzyme on glove fingertips [10].
- Sampling: Directly scrub the fruit surface (e.g., apple, orange) for 10-15 seconds using the sensor-integrated glove fingertip [10].
- Inhibition Reaction: Allow 2-5 minutes for pesticide-enzyme interaction on the glove surface.
- Electrochemical Measurement: Transfer the glove to a portable potentiostat and perform chronoamperometry or square-wave voltammetry measurements in buffer solution containing enzyme substrate.
- Signal Analysis: Quantify the reduction in electrochemical current relative to a pesticide-free control. The signal inhibition is proportional to the pesticide concentration [10].
Performance Characteristics:
- Detection Limit: Nanomolar range (high ppt) for dichlorvos, below EU regulatory limits [10].
- Repeatability: <10% RSD [10].
- Analysis Time: <10 minutes total [10].
Protocol 2: Acetylcholinesterase-Based Inhibition Biosensor for Multi-Pesticide Screening
Principle: This protocol utilizes acetylcholinesterase (AChE) inhibition for detecting organophosphorus and carbamate pesticides in fruit extracts, suitable for laboratory-based high-throughput screening.
Workflow Overview:
Materials and Reagents:
- Acetylcholinesterase (AChE): Primary recognition element from electric eel or recombinant source.
- Acetylthiocholine Iodide: Enzyme substrate.
- Screen-Printed Carbon Electrodes: Disposable electrochemical platforms.
- Phosphate Buffer (0.1 M, pH 7.4): Reaction medium.
- Methanol or Acetonitrile: Extraction solvents.
- C18 Solid-Phase Extraction Cartridges: For sample clean-up.
Procedure:
- Sample Preparation: Homogenize 10 g fruit tissue with 20 mL methanol. Centrifuge at 5000 rpm for 10 minutes. Evaporate supernatant and reconstitute in buffer.
- Extract Clean-up: Pass fruit extract through C18 SPE cartridge to remove interfering compounds.
- Biosensor Assembly: Immobilize AChE on screen-printed carbon electrodes via cross-linking with glutaraldehyde.
- Inhibition Assay: Incubate AChE-modified electrode with 100 μL fruit extract for 10 minutes.
- Electrochemical Measurement: Transfer electrode to buffer containing 1 mM acetylthiocholine. Measure thiocholine oxidation current at +0.45 V vs. Ag/AgCl.
- Quantification: Calculate percentage inhibition relative to pesticide-free control: % Inhibition = [(Icontrol - Isample)/I_control] × 100.
Performance Characteristics:
- Detection Limits: 0.1-5 nM for most organophosphorus pesticides [9].
- Linear Range: 0.5-100 nM.
- Recovery: 85-110% for spiked fruit samples.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Pesticide Residue Biosensor Research
| Reagent/Material | Function/Application | Examples/Specifications |
|---|---|---|
| Acetylcholinesterase (AChE) | Primary biorecognition element for organophosphorus and carbamate detection [9] | Electric eel AChE (Type VI-S), recombinant human AChE |
| Butyrylcholinesterase (BChE) | Broader substrate specificity biorecognition element [10] | Human serum BChE, recombinant expression |
| Screen-Printed Electrodes (SPEs) | Miniaturized, disposable transduction platforms [10] | Carbon, gold, or platinum working electrodes; Ag/AgCl reference |
| Prussian Blue Nanoparticles | High-efficiency electrocatalyst for hydrogen peroxide reduction [10] | Electrochemically synthesized, ~20-50 nm diameter |
| Carbon Black Nanomaterials | Enhanced electron transfer and surface area [10] | Vulcan XC-72, Super P Li |
| Acetylthiocholine Chloride | Enzyme substrate for electrochemical detection [12] | ≥98% purity, electrochemical generation of thiocholine |
| Molecularly Imprinted Polymers (MIPs) | Synthetic biomimetic recognition elements [11] [12] | Methacrylic acid-based polymers imprinted with target pesticides |
| Aptamers | Nucleic acid-based recognition elements [11] [12] | Single-stranded DNA/RNA selected via SELEX process |
| Nanozymes (SAzymes) | Nanomaterial-based enzyme mimics with enhanced stability [12] | Single-atom catalysts (e.g., SACe-N-C) with peroxidase-like activity |
| Portable Potentiostats | Field-deployable electrochemical measurement [10] | PalmSens, EmStat Pico, ADI µPotentiostat |
Data Analysis and Interpretation
Quantitative Risk Assessment Metrics
Table 3: Key Analytical Parameters for Pesticide Risk Assessment Biosensors
| Analytical Parameter | Target Value | Regulatory Significance |
|---|---|---|
| Limit of Detection (LOD) | <1 nM (or lower than MRL) [10] | Enables detection below maximum residue limits (MRLs) |
| Analysis Time | <15 minutes [10] | Suitable for on-site decision making |
| Recovery Percentage | 85-115% [12] | Indicates minimal matrix effects |
| Repeatability (RSD) | <10% [10] | Ensures measurement reliability |
| Linear Dynamic Range | 3 orders of magnitude [12] | Covers sub-MRL to above-MRL concentrations |
Correlation with Health Risk Thresholds
Electrochemical biosensor data must be interpreted within the context of established health risk thresholds:
- Acute Risk Assessment: Compare measured concentrations with Acute Reference Doses (ARfDs) established by regulatory bodies. Sensor signals corresponding to concentrations exceeding 10% of ARfD should trigger immediate risk mitigation.
- Chronic Risk Assessment: For cumulative neurotoxicants, compare detected levels with Acceptable Daily Intakes (ADIs). Consistent detection even below MRLs but above ADI-equivalent levels warrants longitudinal exposure monitoring.
- Cumulative Risk Assessment: For pesticides with common mechanisms of toxicity (e.g., all AChE inhibitors), biosensor signals should be summed to assess aggregate risk, particularly important for multi-residue detection platforms.
Troubleshooting and Technical Notes
- Matrix Effects: Fruit peel matrices (oils, waxes, pigments) can interfere with detection. Always include matrix-matched calibration curves and consider solid-phase extraction for complex matrices [14].
- Enzyme Stability: Cholinesterase enzymes can denature under field conditions. Implement proper storage (lyophilized forms) and consider nanozymes or MIPs for improved stability [12].
- Cross-Reactivity: AChE-based biosensors cannot distinguish between different organophosphorus or carbamate pesticides. Confirm positive results with specific methods if compound identification is required [9].
- Sensor Regeneration: For reversible inhibitors (carbamates), sensors can be regenerated with oxime solutions (e.g., pralidoxime). Irreversible inhibitors (organophosphates) typically require sensor replacement [9].
Electrochemical biosensor technology continues to evolve toward multi-analyte detection, artificial intelligence-enhanced data processing, and integration with wireless connectivity for real-time food safety monitoring [11] [14]. These advances will further strengthen the correlation between detected residue levels and their potential health impacts, enabling more precise risk assessment and management across the food supply chain.
The One Health approach is defined as "a collaborative, multisectoral, and transdisciplinary approach — working at the local, regional, national, and global levels — with the goal of achieving optimal health outcomes recognizing the interconnection between people, animals, plants, and their shared environment" [15]. This perspective is particularly crucial for addressing the challenge of pesticide residues in food, a quintessential One Health issue that sits at the intersection of agricultural practices, environmental health, and human well-being [16] [15].
Foodborne diseases (FBDs), which can result from pesticide contamination, impose a significant global burden, causing over 100 million USD in annual preventable economic losses, with over 90% of this burden affecting low- and middle-income countries (LMICs) [16]. These diseases disproportionately impact children under five years of age, who experience 38% of all FBD incidence despite representing only 9% of the global population [16]. The interconnected issues of dwindling animal and plant health, food systems vulnerable to contamination, and pathogen threats necessitate a unified framework that concurrently addresses the health of humans, animals, and ecosystems [16].
Electrochemical biosensors have emerged as powerful tools within this framework, enabling the sensitive detection of pesticide residues in food matrices. These devices complement traditional chromatography methods like HPLC and GC-MS, which though highly accurate, require expensive equipment, extensive sample pretreatment, and highly skilled professionals [17]. Biosensors offer a viable alternative that simplifies or removes complex preparation steps, providing rapid, on-site analysis capabilities essential for monitoring the food supply chain [17].
Analytical Performance of Nanomaterial-Based Biosensors for Pesticide Detection
The integration of nanomaterials into biosensing platforms has significantly enhanced their analytical performance. The table below summarizes the detection capabilities of various nanomaterial-based biosensors for specific pesticides in food matrices, demonstrating limits of detection (LOD) well below the Codex Alimentarius maximum residue limits [17].
Table 1: Analytical Performance of Nanomaterial-Based Biosensors for Pesticide Detection in Food
| Nanomaterial | Biorecognition Element | Pesticide Detected | Limit of Detection (LOD) | Food Matrix | Transducer Type |
|---|---|---|---|---|---|
| Gold Nanoparticles (AuNPs) | Acetylcholinesterase (AChE) | Organophosphorus (11 types) | 19–77 ng L⁻¹ | Apple, Cabbage | Electrochemical |
| Gold Nanoparticles (AuNPs) | Acetylcholinesterase (AChE) | Methomyl | 81 ng L⁻¹ | Apple, Cabbage | Electrochemical |
| Gold Nanoparticles (AuNPs) | Aptamer | Chlorpyrifos | 36 ng L⁻¹ | Apple, Pak choi | Electrochemical |
| Gold Nanoparticles (AuNPs) | Antibody | Chlorpyrifos | 0.07 ng L⁻¹ | Chinese cabbage, Lettuce | Electrochemical |
| Gold Nanoparticles (AuNPs) | AChE | Carbamate | 1.0 nM | Fruit | Electrochemical |
| Nanohybrids | Various | Various | Varies (typically < 100 ng L⁻¹) | Various fruits, vegetables | Electrochemical, Fluorescent |
The data reveals that electrochemical transducers are the most prevalent (71.18% of studies), followed by fluorescent (13.55%) and colorimetric (8.47%) systems [17]. The exceptional sensitivity of these platforms, particularly those utilizing noble metal nanoparticles and carbon-based nanomaterials, enables detection at picomolar levels, ensuring food safety even for trace contaminants [17].
Research Reagent Solutions: Essential Materials for Biosensor Development
The development of high-performance biosensors requires carefully selected materials and reagents. The following table details key components and their functions in constructing electrochemical biosensors for pesticide detection.
Table 2: Essential Research Reagents and Materials for Biosensor Construction
| Reagent/Material | Function/Application | Examples & Notes |
|---|---|---|
| Nanomaterials | Enhance sensitivity, conductivity, and catalytic activity; provide high surface area for bioreceptor immobilization | AuNPs, AgNPs, Carbon NDs, MWCNTs, Nanohybrids [17] |
| Biorecognition Elements | Provide selective binding to target pesticide molecules | AChE enzyme, Aptamers, Antibodies, MIPs [17] |
| Transducer Materials | Convert biological recognition event into measurable electrical signal | Screen-printed electrodes (SPCE, SPWPE), Glassy Carbon Electrode (GCE) [17] |
| Chemical Reagents | Facilitate electrode modification, signal amplification, and experimental procedures | Tri-n-propylamine (TprA), Bovine Serum Albumin (BSA), Glutaraldehyde (for cross-linking) [17] |
| Buffer Solutions | Maintain optimal pH and ionic strength for biological components | Phosphate buffer saline (PBS) for enzyme stability and binding reactions |
Experimental Protocol: Development of an Acetylcholinesterase-Based Electrochemical Biosensor
Apparatus and Reagents
- Electrochemical Workstation: Potentiostat with capability for cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS)
- Electrodes: Screen-printed carbon electrodes (SPCEs) or glassy carbon electrodes (GCE)
- Nanomaterials: Gold nanoparticles (AuNPs, 20nm diameter) and multi-walled carbon nanotubes (MWCNTs)
- Biological Reagents: Acetylcholinesterase (AChE) enzyme, acetylthiocholine iodide (ATCh) substrate
- Buffer Solutions: Phosphate buffer saline (PBS, 0.1M, pH 7.4)
- Pesticide Standards: Chlorpyrifos, malathion, and paraoxon stock solutions (1000 ppm in methanol)
Electrode Modification and Biosensor Fabrication
- Electrode Pretreatment: Clean GCE with alumina slurry and perform cyclic voltammetry scans in 0.5M H₂SO₄ from -0.2V to +1.5V until stable response achieved.
- Nanomaterial Deposition: Prepare nanohybrid suspension (1mg/mL AuNPs-MWCNTs in DMF) and drop-cast 10μL onto electrode surface. Dry at room temperature for 2 hours.
- Enzyme Immobilization: Incubate modified electrode with 10μL AChE solution (0.5U/μL) in PBS at 4°C for 12 hours.
- Stabilization: Rinse electrode with PBS to remove unbound enzyme and stabilize in PBS for 1 hour before use.
Pesticide Detection Procedure
- Baseline Measurement: Record CV or EIS response in PBS containing 1mM ATCh as substrate.
- Inhibition Assay: Incubate biosensor with pesticide standard or sample solution for 10 minutes.
- Measurement: Record electrochemical response after incubation under identical conditions to baseline.
- Quantification: Calculate inhibition percentage using formula: % Inhibition = [(I₀ - I₁)/I₀] × 100 where I₀ is current before inhibition and I₁ is current after inhibition.
- Calibration: Generate standard curve using known pesticide concentrations and interpolate sample values.
Sample Preparation (Fruit Matrices)
- Extraction: Homogenize 10g fruit sample with 20mL acetonitrile in a blender for 2 minutes.
- Cleanup: Filter through Buchner funnel and concentrate extract under nitrogen stream.
- Reconstitution: Dilute concentrate 1:10 with PBS for analysis.
- Recovery Test: Perform standard addition method to validate accuracy.
Workflow Visualization: One Health Perspective in Pesticide Monitoring
Diagram 1: One Health pesticide monitoring workflow.
Biosensor Signaling Pathway for Pesticide Detection
Diagram 2: Biosensor signaling pathway with inhibition mechanism.
Data Analysis and Interpretation Guidelines
Calibration and Quantification
- Plot inhibition percentage versus pesticide concentration on a semi-log scale to generate calibration curves
- Determine linear range and limit of detection (LOD) using 3σ/slope criterion
- Calculate limit of quantification (LOQ) using 10σ/slope criterion
- For real samples, apply standard addition method to account for matrix effects
Validation Parameters
- Accuracy: Evaluate through recovery studies (85-115% acceptable range)
- Precision: Determine through relative standard deviation (RSD < 10% for replicates)
- Selectivity: Test against common interferents (heavy metals, other pesticides)
- Stability: Monitor biosensor response over 30-day period with proper storage
Comparison with Regulatory Standards
- Compare detected pesticide levels with Codex Alimentarius Maximum Residue Limits (MRLs)
- Reference WHO guidelines for acceptable daily intake (ADI) values
- Consider cumulative risk assessment for multiple pesticide residues
Electrochemical biosensors represent a transformative technology for operationalizing the One Health approach to pesticide monitoring. Their ability to provide rapid, sensitive, and on-site detection of harmful residues directly connects agricultural practices with human health outcomes through the shared environment [16] [15] [17]. The protocols outlined herein provide researchers with robust methodologies for developing these analytical tools, contributing to the broader goal of reducing the burden of foodborne diseases and promoting sustainable agricultural systems that respect the interconnected health of people, animals, and ecosystems.
The accurate detection of pesticide residues on fruits is a critical component of food safety monitoring, essential for protecting public health. For decades, the field has been dominated by conventional analytical techniques, primarily chromatography-based methods. While these methods are recognized for their accuracy and sensitivity, they possess significant drawbacks that limit their practical application for rapid, on-site screening. This document details the specific limitations of these conventional methods, focusing on their high cost, operational complexity, and lack of portability. Furthermore, it positions electrochemical biosensors as a promising alternative, outlining their working principles and advantages to provide researchers with a clear rationale for the paradigm shift in pesticide detection protocols.
Quantitative Analysis of Conventional Method Limitations
Conventional techniques for pesticide residue analysis, such as gas chromatography-tandem mass spectrometry (GC-MS/MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS), while highly accurate, present substantial barriers to widespread and efficient use [18]. The following table summarizes the core limitations of these established methods.
Table 1: Core Limitations of Conventional Pesticide Detection Methods
| Limitation | Key Characteristics | Impact on Research & Deployment |
|---|---|---|
| High Cost [18] | - Significant initial capital investment for instruments (chromatographs, mass spectrometers).- Ongoing expenses for high-purity gases, solvents, and consumables.- Requirement for specialized laboratory infrastructure and maintenance. | Prohibits adoption in resource-limited settings, including field stations and smaller laboratories. Increases the per-sample cost of analysis. |
| Operational Complexity [18] | - Requires multi-step sample preparation (extraction, clean-up, pre-concentration).- Necessitates highly trained, specialized personnel for operation and data interpretation.- Time-consuming procedures, limiting sample throughput and delaying results. | Creates a bottleneck for high-volume screening. Results in a dependency on expert operators, increasing labor costs and limiting scalability. |
| Lack of Portability [18] | - Instruments are large, heavy, and require a stable laboratory environment.- Not suitable for on-site, at-line, or point-of-care testing at farms, markets, or border inspections. | Prevents real-time decision-making and rapid intervention. Requires sample transport, which can compromise integrity and increase time-to-result. |
In addition to these core limitations, traditional biorecognition elements like enzymes and antibodies, used in some sensors, can suffer from instability and complex preparation requirements [19]. The development of aptamer-based sensors (aptasensors) has emerged to overcome these issues, offering superior stability, reusability, and simpler production [19].
Experimental Protocols for Comparison
To illustrate the contrast between conventional and emerging methods, the following protocols detail a standard laboratory-based analysis versus a novel, portable biosensor approach.
Protocol A: Conventional LC-MS/MS Analysis for Multi-Pesticide Residues
This protocol is adapted from established methods for determining pesticide residues in complex food matrices like fruits [18].
1. Principle: Pesticides are extracted from a homogenized fruit sample, purified to remove interfering compounds, separated via liquid chromatography, and then identified and quantified by tandem mass spectrometry based on their mass-to-charge ratio.
2. Materials and Reagents:
- Homogenized fruit sample (e.g., apple, orange peel)
- Organic solvents (Acetonitrile, Methanol) of HPLC grade
- QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) extraction kits
- Centrifuge tubes and vials
- High-performance liquid chromatography system coupled to a tandem mass spectrometer (LC-MS/MS)
- Analytical column (e.g., C18 reverse-phase)
3. Procedure: 1. Sample Preparation: Weigh 10 g of homogenized fruit sample into a 50 mL centrifuge tube. 2. Extraction: Add 10 mL of acetonitrile and shake vigorously for 1 minute. Use a QuEChERS salt packet to induce phase separation and centrifuge. 3. Clean-up: Transfer an aliquot of the upper acetonitrile layer to a dispersive Solid-Phase Extraction (d-SPE) tube containing sorbents. Shake and centrifuge to remove impurities. 4. Pre-concentration: Evaporate a portion of the clean extract to dryness under a gentle nitrogen stream and reconstitute in a smaller volume of initial mobile phase. 5. Instrumental Analysis: - Chromatographic Separation: Inject the reconstituted extract into the LC system. Pesticides are separated as they travel through the column under a specific gradient of aqueous and organic mobile phases. - MS Detection: Eluting compounds are ionized and fragmented in the mass spectrometer. Detection is based on monitoring unique precursor-product ion transitions for each pesticide. 6. Data Analysis: Quantify pesticide concentrations by comparing the peak areas of samples to those of a calibrated standard curve.
Protocol B: On-Glove Electrochemical Biosensor for Organophosphorus Pesticides
This protocol details a decentralized analysis method using a biosensor integrated directly onto a glove for sampling and detection on fruit peels [10].
1. Principle: A glove is fitted with an electrochemical biosensor containing the enzyme butyrylcholinesterase. Organophosphorus pesticides (e.g., dichlorvos) inhibit this enzyme. The degree of inhibition, measured via a change in electrochemical current, is proportional to the pesticide concentration.
2. Materials and Reagents:
- Glove with an integrated screen-printed electrode (SPE)
- Biosensor modification components: Prussian blue, Carbon black, Butyrylcholinesterase enzyme
- Portable potentiostat for electrochemical measurements
- Buffer solution
3. Procedure: 1. Sampling: The user wears the modified glove and simply scrubs the surface of the fruit (e.g., an apple) with the finger containing the biosensor. This direct contact transfers the pesticide residue from the peel to the sensor. 2. Measurement: The user places the sensor-finger into the portable potentiostat to perform an electrochemical measurement (e.g., chronoamperometry). 3. Detection: The system measures the enzymatic activity. A significant reduction in the current signal compared to a baseline indicates the presence of enzyme-inhibiting pesticides. 4. Analysis: The concentration of pesticide is quantified from the measured current using a pre-calibrated curve. The entire process, from sampling to result, is completed on-site in minutes.
Visualizing Workflows and Advantages
The fundamental differences in the workflows and capabilities of conventional methods versus portable biosensors are visualized below.
Diagram 1: A comparison of analytical workflows, highlighting the streamlined process of biosensors.
The Scientist's Toolkit: Research Reagent Solutions
The development and operation of advanced electrochemical biosensors rely on key materials and reagents. The following table details essential components for constructing and using an on-glove biosensor for pesticide detection, as featured in the cited protocol [10].
Table 2: Essential Research Reagents for an On-Glove Electrochemical Biosensor
| Item | Function/Description | Application Note |
|---|---|---|
| Butyrylcholinesterase Enzyme | The biological recognition element that specifically reacts with its substrate; its activity is inhibited by organophosphorus pesticides. | The core of the biosensor's specificity. Requires stable immobilization on the electrode surface to maintain activity. |
| Screen-Printed Electrode (SPE) | A disposable, miniaturized electrochemical cell (working, counter, and reference electrodes) printed on a plastic or ceramic substrate. | Provides the platform for the biosensor. Ideal for mass production and integration into wearable devices like gloves. |
| Prussian Blue (PB) | An electron transfer mediator that shuttles electrons between the enzyme and the electrode, enhancing the current signal. | Often called an "artificial peroxidase," it improves the sensitivity and lowers the operating potential of the sensor. |
| Carbon Black | A nanomaterial used to modify the electrode surface, increasing its effective surface area and improving electron conductivity. | Enhances the electrochemical signal and provides a robust matrix for immobilizing the enzyme and mediator. |
| Portable Potentiostat | A compact, battery-powered electronic instrument that applies potential and measures the resulting current in an electrochemical cell. | Enables on-site and real-time measurements. Critical for moving analysis out of the centralized laboratory. |
Definition and Core Principle
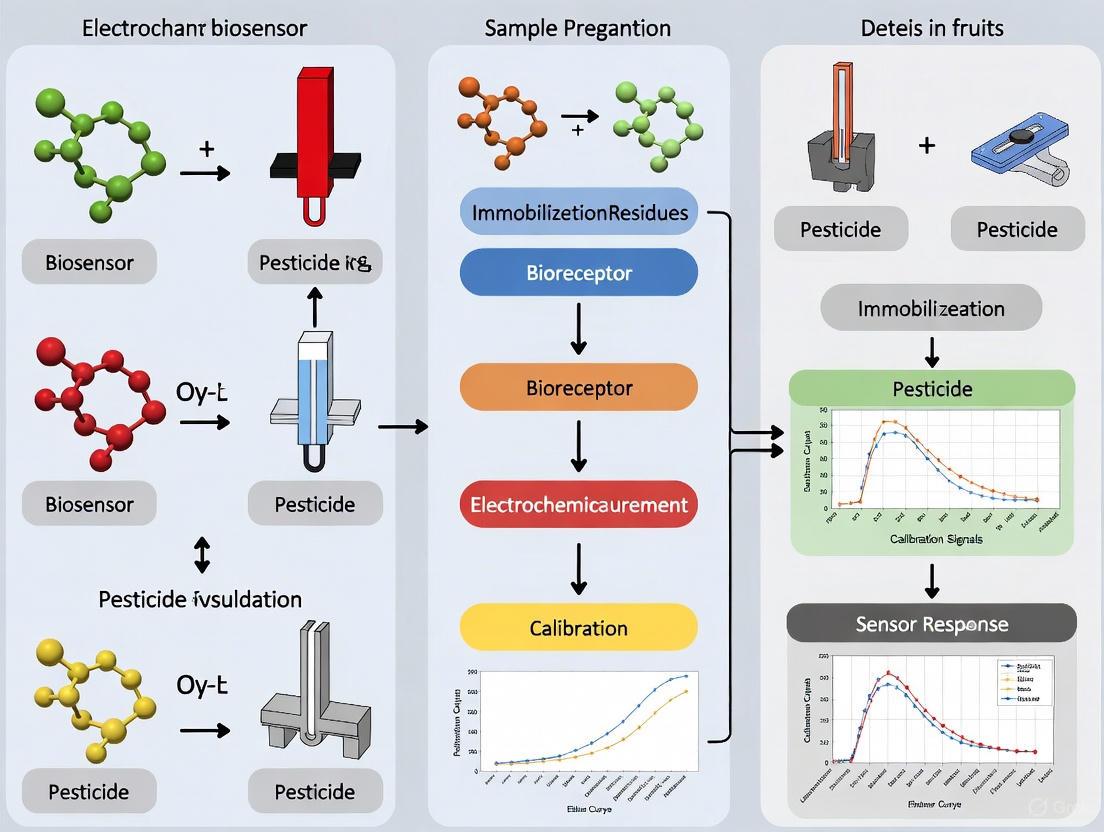
An electrochemical biosensor is defined as a self-contained integrated device that converts a biological response into a quantifiable and processable electronic signal [20] [21]. These sensors utilize a biological recognition element (such as an enzyme, antibody, or nucleic acid) that is retained in direct spatial contact with an electrochemical transduction element [21]. The core principle involves the direct conversion of a biological event—such as an enzyme-substrate reaction or an antigen-antibody interaction—into an electrical signal (e.g., current, voltage, or impedance) [22]. This distinguishes true biosensors from bioanalytical systems that require additional processing steps like reagent addition [21].
Fundamental Components and Architecture
Electrochemical biosensors consist of five main components that work in sequence to detect and report analytical information [20] [23].
- Bioreceptor: This biological recognition element (e.g., enzyme, antibody, DNA, or whole cell) selectively binds to the target analyte. The specificity of the bioreceptor determines the sensor's selectivity [20] [22].
- Interface Architecture: This is the physical space where the specific biological event (e.g., binding or catalysis) occurs. The interface is often engineered with nanomaterials to enhance performance [20] [22].
- Transducer Element: Typically an electrode, this component transforms the biochemical signal resulting from the bioreceptor-analyte interaction into a measurable electrical signal [20] [23].
- Signal Processor: This detector circuit converts and amplifies the transducer's signal into an electronic signal that can be processed [20] [23].
- User Interface: Computer software and a display convert the electronic signal into a meaningful physical parameter (e.g., analyte concentration) that is presented to the operator [20] [23].
The performance of an electrochemical biosensor is heavily influenced by the surface architectures at the nanoscale that connect the sensing element to the biological sample, affecting both signal transduction and overall sensitivity [20] [22].
Classification of Electrochemical Biosensors
Electrochemical biosensors can be classified based on their transduction method and the type of biological recognition element used [24] [21]. The most common classification is by transduction principle, as detailed in the table below.
Table 1: Classification of Electrochemical Biosensors by Transduction Principle
| Transducer Type | Measured Parameter | Principle of Operation | Key Advantages | Example Application in Pesticide Detection |
|---|---|---|---|---|
| Amperometric [24] [25] | Current | Measures current from electrochemical oxidation/reduction of an electroactive species at a constant applied potential [24]. | High sensitivity, rapid response [24]. | Detection of organophosphorus pesticides using acetylcholinesterase inhibition [17]. |
| Voltammetric [24] | Current | Similar to amperometric, but the applied potential is ramped (e.g., cyclic or differential pulse voltammetry) and the resulting current is measured [24]. | Provides quantitative and qualitative data [24]. | Detection of chlorpyrifos using aptamer-based sensors on gold nanoparticles (LOD: 36 ng L⁻¹) [17]. |
| Potentiometric [24] [23] | Potential (Voltage) | Measures the accumulation of charge at an electrode (vs. a reference electrode) at zero current flow [24] [25]. | Small size, rapid response, resistant to color/turbidity [24]. | Often used with ion-selective electrodes for ion detection [25]. |
| Impedimetric [24] [25] | Impedance | Measures resistive and capacitive changes in the system by applying a small-amplitude AC potential. Can be label-free [24]. | Label-free, real-time monitoring of binding events [24]. | Label-free immunosensing for detection of dengue virus protein [24]. |
| Field-Effect Transistor (FET) [24] [26] | Current/Conductivity | Detects changes in source-drain channel conductivity caused by charged target species accumulating at the sensor surface [24]. | Label-free, miniaturization, mass production potential [24]. | Highly sensitive detection of Lyme disease antigens (LOD: 2×10⁻³ ng mL⁻¹) [24]. |
Operational Advantages
Electrochemical biosensors offer a compelling set of advantages that make them particularly suitable for on-site analysis, including the detection of pesticide residues in fruit [20] [23] [17].
- High Sensitivity and Low Detection Limits: These sensors can achieve excellent detection limits, often down to picomolar concentrations, due to the efficient transduction of biochemical events into electrical signals [25]. For example, biosensors for pesticides like chlorpyrifos have demonstrated limits of detection (LOD) as low as 70 × 10⁻³ ng L⁻¹, which is well below the maximum residue limits set by regulatory bodies [17].
- Instrumental Simplicity and Low Cost: They do not require complex or expensive optical components. Their design benefits from advances in low-cost microelectronic production, making the equipment affordable and easy to interface with standard readout systems [24] [23].
- Portability and Ease of Miniaturization: The electrochemical platform is inherently suitable for creating small, portable, and handheld devices, which is ideal for field-use and point-of-care testing [20] [23].
- Ability to Analyze Turbid Samples and Small Volumes: Unlike many optical methods, electrochemical sensors can function effectively in turbid biofluids and require only small volumes of analyte for analysis [20] [23].
- Rapid Response and Real-Time Analysis: These biosensors can provide results in minutes, allowing for real-time or near-real-time monitoring of analytes, which is crucial for quick decision-making in food safety [20] [17].
Experimental Protocol: Amperometric Biosensor for Organophosphorus Pesticide Detection
This protocol outlines the development and use of an amperometric biosensor for detecting organophosphorus pesticides in fruit samples, based on the inhibition of the enzyme acetylcholinesterase (AChE) [17].
Principle
Organophosphorus pesticides inhibit the activity of AChE. The biosensor measures the reduction in enzymatic activity by monitoring the amperometric current generated from the enzymatic reaction of AChE with its substrate, acetylthiocholine. The decrease in current is proportional to the pesticide concentration [17].
Materials and Reagents
Table 2: Research Reagent Solutions and Essential Materials
| Item | Function/Description | Specifications/Notes |
|---|---|---|
| Acetylcholinesterase (AChE) [17] | Biorecognition element; catalyzes substrate reaction. | From electric eel or recombinant source; immobilizable. |
| Acetylthiocholine [17] | Enzyme substrate; produces electroactive product upon hydrolysis. | Alternative to acetylcholine for more stable measurement. |
| Gold Nanoparticles (AuNPs) [17] [22] | Nanomaterial for electrode modification; increases surface area and enhances electron transfer. | ~10-20 nm diameter; can be synthesized or commercially acquired. |
| Screen-Printed Carbon Electrode (SPCE) [17] | Disposable electrochemical cell (working, counter, and reference electrodes). | Enables portability and single-use applications. |
| Phosphate Buffered Saline (PBS) | Electrolyte solution; provides optimal pH and ionic strength for enzymatic activity. | Typically 0.1 M, pH 7.4. |
| Fruit Sample Extract | Test matrix; requires pre-processing (blending, filtration, dilution). | Apple, cabbage, and other fruits have been successfully tested [17]. |
Step-by-Step Procedure
Step 1: Electrode Modification and Enzyme Immobilization
- Prepare the working electrode: Clean the surface of the screen-printed carbon electrode (SPCE) according to manufacturer's instructions.
- Apply nanomaterial: Drop-cast 5-10 µL of a suspension of Gold Nanoparticles (AuNPs) onto the working electrode surface and allow it to dry at room temperature. This step increases the active surface area and improves electron transfer [17] [22].
- Immobilize the enzyme: Apply 5 µL of the AChE enzyme solution (e.g., 100 mU/µL in PBS) onto the AuNP-modified SPCE. Incubate in a humid chamber for 1 hour at 4°C to allow for physical adsorption or covalent binding.
- Rinse: Gently rinse the modified electrode with PBS buffer to remove any unbound enzyme.
Step 2: Apparatus Setup and Baseline Measurement
- Connect the biosensor: Place the modified SPCE into the potentiostat and connect the electrical contacts.
- Add electrolyte: Pipette 50 µL of PBS into the electrochemical cell.
- Measure baseline current: Add 10 µL of acetylthiocholine substrate solution (final concentration 1 mM) to the cell. Immediately apply a constant working potential of +0.5 V (vs. the Ag/AgCl reference electrode) and record the steady-state current (i_baseline). This current is generated by the oxidation of thiocholine, the product of the enzymatic hydrolysis of acetylthiocholine.
Step 3: Inhibition (Pesticide Detection)
- Expose to sample: Incubate a separate, identical AChE/AuNP/SPCE biosensor in 50 µL of the prepared fruit sample extract for 10 minutes.
- Rinse: Rinse the electrode gently with PBS to remove the sample while retaining the inhibited enzyme.
- Measure sample current: Add 10 µL of acetylthiocholine substrate as in Step 2.3 and record the steady-state current (i_sample).
Step 4: Data Analysis
- Calculate Inhibition: The percentage of enzyme inhibition is calculated as follows:
Inhibition (%) = [(i_baseline - i_sample) / i_baseline] * 100 - Quantify Pesticide: The percentage inhibition is correlated to the pesticide concentration by interpolating from a calibration curve previously constructed using standard solutions of known pesticide concentrations.
The following diagram illustrates the experimental workflow:
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Electrochemical Biosensor Research in Pesticide Detection
| Category | Item | Function in Experiment |
|---|---|---|
| Biorecognition Elements | Acetylcholinesterase (AChE) [17] | Key enzyme for organophosphate/carbamate detection; inhibition is measured. |
| Nucleic Acid Aptamers [17] | Synthetic single-stranded DNA/RNA molecules that bind specific pesticides (e.g., chlorpyrifos). | |
| Monoclonal Antibodies [17] | Provide high specificity for immunoassays; used in immunosensors. | |
| Nanomaterials | Gold Nanoparticles (AuNPs) [17] [22] | Enhance electron transfer and provide a large surface area for biomolecule immobilization. |
| Carbon Nanotubes (SWCNTs/MWCNTs) [22] | Improve conductivity and catalytic activity; used to modify electrode surfaces. | |
| Graphene Oxide / Reduced GO [22] | 2D carbon material with high surface area and good dispersibility for sensor fabrication. | |
| Electrode & Instrumentation | Screen-Printed Electrodes (SPEs) [17] | Low-cost, disposable, and portable electrochemical cells ideal for on-site testing. |
| Potentiostat/Galvanostat | Core instrument for applying potential and measuring current in amperometric/voltammetric sensors. | |
| Supporting Reagents | Redox Probes (e.g., [Fe(CN)₆]³⁻/⁴⁻) [24] | Used in impedimetric and voltammetric sensors to facilitate electron transfer and measure changes. |
| Blocking Agents (e.g., BSA) [17] | Used to cover non-specific binding sites on the sensor surface to reduce background noise. |
A Step-by-Step Protocol: From Biosensor Fabrication to On-Site Fruit Analysis
The accurate monitoring of pesticide residues in fruits is paramount for ensuring global food safety. Electrochemical biosensors have emerged as powerful analytical tools for this purpose, combining high sensitivity with the potential for rapid, on-site analysis. The performance of these biosensors is fundamentally governed by the biorecognition element (BRE) immobilized on the transducer surface, which dictates the sensor's selectivity, sensitivity, and operational stability [27]. This application note provides a detailed comparison of four principal classes of BREs—enzymes, aptamers, antibodies, and molecularly imprinted polymers (MIPs)—within the context of developing robust electrochemical biosensors for fruit pesticide residue analysis. We summarize their characteristics in structured tables and provide detailed experimental protocols to guide researchers in the selection, optimization, and application of these critical components.
Comparative Analysis of Biorecognition Elements
The selection of a BRE involves balancing factors such as specificity, stability, cost, and ease of fabrication. Table 1 provides a quantitative comparison of these elements, while Table 2 outlines their suitability for detecting different pesticide classes.
Table 1: Performance Comparison of Biorecognition Elements for Pesticide Biosensors
| Biorecognition Element | Affinity & Sensitivity | Stability & Lifetime | Development Cost & Time | Key Advantages | Primary Limitations |
|---|---|---|---|---|---|
| Enzymes | Moderate sensitivity; operates on inhibition principle [28] | Low; susceptible to denaturation, short lifetime [27] | Low to moderate cost; readily available [28] | "Biologically relevant" detection mechanism; reusable after reactivation [28] | Limited to pesticides that are enzyme inhibitors; susceptible to environmental conditions [27] |
| Aptamers | High affinity; detection limits down to femtomolar (fM) range [19] | High; stable over long-term storage and tolerant to harsh conditions [19] [27] | Moderate SELEX cost; inexpensive in vitro synthesis [19] | Small size, high stability, reusable, amenable to chemical modification [19] [27] | Susceptible to nuclease degradation in some environments; complex SELEX process for new targets [19] |
| Antibodies | Very high affinity and specificity [27] | Moderate; sensitive to temperature and pH [27] | High cost and time for development and production [27] | Well-established, high specificity validation protocols [27] | Animal-derived production; batch-to-batch variation; irreversible binding [19] [27] |
| Molecularly Imprinted Polymers (MIPs) | High selectivity; comparable to antibodies ("artificial antibodies") [29] | Very high; robust thermal and chemical stability [29] | Low cost, rapid synthesis [29] | Excellent physical/chemical stability; reusable; suitable for harsh environments [27] [29] | Occasional incomplete template removal; heterogeneous binding sites [27] |
Table 2: Biorecognition Element Suitability for Major Pesticide Classes
| Pesticide Class | Example Pesticides | Suitable Biorecognition Elements | Detection Mechanism Notes |
|---|---|---|---|
| Organophosphates (OPs) | Dichlorvos, Malathion, Parathion [28] [10] | Enzymes (Cholinesterases), Aptamers, Antibodies, MIPs | Enzymatic inhibition is dominant for OPs and carbamates [28] [27]. Aptamers/MIPs allow specific compound identification [30]. |
| Carbamates | Carbofuran, Carbaryl, Aldicarb [28] | Enzymes (Cholinesterases), Aptamers, MIPs | Same neurotoxic mechanism as OPs allows use of same enzyme-based sensors [28]. |
| Triazines & Phenylureas | Atrazine, Diuron | Aptamers, Antibodies, MIPs, Photosynthetic Enzymes (e.g., PSII) | Detection often relies on direct binding. Photosystem II inhibition is a specific mechanism for herbicides [28]. |
| Organochlorines (OCPs) | DDT, Lindane | Antibodies, Aptamers, MIPs | Typically detected via direct binding assays due to their environmental persistence [18]. |
| Neonicotinoids | Thiamethoxam, Imidacloprid | Aptamers, Antibodies [19] | Direct binding is the primary mode of detection for these systemic insecticides [19]. |
The following diagram illustrates the strategic decision-making workflow for selecting the optimal biorecognition element based on research objectives and practical constraints.
Detailed Experimental Protocols
Protocol 1: Fabrication of an On-Glove Enzyme-Based Biosensor
This protocol details the construction of an innovative on-glove biosensor for the direct detection of organophosphorus pesticides (e.g., dichlorvos) on fruit peels, enabling decentralized analysis [10].
1. Reagents and Materials:
- Screen-printed carbon electrode (SPCE)
- Butyrylcholinesterase (BChE) enzyme
- Prussian blue (PB) and Carbon black (CB) nanoparticles
- Glove (e.g., nitrile)
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
- Acetylthiocholine iodide (ATCh) substrate
- Dichlorvos standard
2. Sensor Fabrication Steps:
- Step 1: Preparation of Bio-hybrid Ink. Disperse Prussian blue and Carbon black nanoparticles in a suitable binder solution (e.g., Nafion) to form a stable carbon ink. Subsequently, mix the carbon ink with a solution of BChE enzyme to create a homogeneous bio-hybrid probe.
- Step 2: Integration onto Glove. Using a precision pipette, deposit a small volume (e.g., 5-10 µL) of the bio-hybrid ink onto the index finger of the glove and allow it to dry at room temperature. The SPCE is affixed to the glove's fingertip for connection to the potentiostat.
- Step 3: Electrochemical Characterization. Characterize the modified electrode using cyclic voltammetry (CV) in PBS to confirm the successful immobilization of the Prussian blue mediator and the enzyme.
3. Sample Analysis Workflow:
- Step 1: Sampling. The user scrubs the surface of the fruit (e.g., apple, orange) with the biosensor-integrated fingertip.
- Step 2: Inhibition. Any OPs present on the fruit peel will inhibit the BChE enzyme on the glove.
- Step 3: Measurement. The user adds a drop of ATCh substrate solution onto the sensor. The enzymatic conversion of ATCh produces thiocholine, which is electrochemically oxidized at the Prussian blue-modified electrode. The measured current is inversely proportional to the pesticide concentration due to the inhibition of BChE.
- Step 4: Quantification. The dichlorvos concentration is calculated from the percentage of enzyme inhibition using a pre-established calibration curve.
Protocol 2: Development of an Electrochemical Aptasensor for Carbendazim
This protocol outlines the construction of a highly sensitive aptasensor for the fungicide carbendazim (CBZ) using a dual-aptamer design and nanomaterial-enhanced signal amplification [19].
1. Reagents and Materials:
- Gold nanoparticles (Au NPs)
- Boron-doped electrode or SPCE
- CBZ-specific aptamer (CBZA) and its complementary strand (SH-cCBZA), thiol-modified
- Zirconium-based Metal-Organic Framework (MOF-808)
- Graphene nanoribbons
- Methylene blue (MB) redox reporter
- Carbendazim standard
2. Sensor Fabrication Steps:
- Step 1: Electrode Modification. Prepare a nanocomposite of graphene nanoribbons and MOF-808. Drop-cast this composite onto the electrode surface to create a high-surface-area platform. Electrodeposit Au NPs onto the modified electrode to facilitate subsequent aptamer immobilization via Au–S bonds.
- Step 2: Aptamer Immobilization. Co-immobilize the thiolated complementary DNA (SH-cCBZA) and the MB-labeled CBZ aptamer (CBZA) onto the Au NP surface. The two strands hybridize, forming a rigid double-stranded DNA (dsDNA) structure on the electrode.
3. Measurement and Detection Principle:
- Principle: In the absence of CBZ, the dsDNA structure keeps the MB reporter in a specific conformation, yielding a baseline electrochemical current.
- Step 1: Incubation. Incubate the aptasensor with the sample solution.
- Step 2: Binding-Induced Displacement. Upon introduction of CBZ, the aptamer has a stronger affinity for the target than for its complementary strand. It undergoes a conformational change, dissociates from the complementary strand, and binds to CBZ. This displacement leads to the removal of the MB-labeled complex from the electrode surface.
- Step 3: Signal Measurement. The change in the MB oxidation current (typically an increase) is measured using differential pulse voltammetry (DPV). The signal change is directly proportional to the CBZ concentration, achieving ultra-trace level detection.
Protocol 3: Construction of a MIP-Based Electrochemical Nanosensor
This protocol describes the creation of a robust and selective sensor using a Molecularly Imprinted Polymer as an artificial antibody for pesticide detection [29].
1. Reagents and Materials:
- Target pesticide molecule (template, e.g., chlorpyrifos)
- Functional monomers (e.g., acrylamide, pyrrole, o-phenylenediamine)
- Cross-linker (e.g., ethylene glycol dimethacrylate - EGDMA)
- Initiator (e.g., ammonium persulfate)
- Solvent (acetonitrile or water)
- Electrode (e.g., glassy carbon, SPCE)
2. Sensor Fabrication Steps:
- Step 1: Pre-Assembly. Mix the template molecule (pesticide) with the functional monomers in a solvent. Allow them to form a complex via non-covalent interactions (e.g., hydrogen bonding, van der Waals forces).
- Step 2: Electropolymerization. Place the electrode in the pre-assembled mixture. Using cyclic voltammetry (CV), apply a potential scan over a defined range to initiate the electrochemical polymerization of the monomers around the template. This process forms a thin, rigid polymer film on the electrode surface with cavities complementary in size, shape, and functionality to the target pesticide.
- Step 3: Template Removal. Thoroughly wash the electrode with a suitable solvent (e.g., methanol:acetic acid mixture) to extract the template molecules from the polymer matrix. This leaves behind specific recognition sites.
3. Measurement and Detection:
- Step 1: Rebinding. Incubate the MIP-modified electrode in the sample solution containing the target pesticide. The pesticide molecules selectively rebind to the imprinted cavities.
- Step 2: Electrochemical Readout. Measure the electrode's response using a technique such as differential pulse voltammetry (DPV) or electrochemical impedance spectroscopy (EIS). The binding of the target pesticide hinders electron transfer, leading to a measurable change in current or impedance, which is correlated with the pesticide concentration.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for Biosensor Fabrication
| Reagent/Material | Function/Application | Key Features & Considerations |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, portable transducer platform; ideal for on-site testing [10] | Low-cost, mass-producible, integrated 3-electrode system (working, reference, counter) |
| Gold Nanoparticles (Au NPs) | Signal amplification; platform for immobilizing thiolated bioreceptors (aptamers, antibodies) [19] | High conductivity, large surface area, biocompatibility, facile surface chemistry (Au–S bonds) |
| Prussian Blue (PB) | Electron transfer mediator in enzyme-based sensors [10] | High electrocatalytic activity for H₂O₂ reduction, low working potential, "artificial peroxidase" |
| Graphene & Derivatives | Electrode nanomodifier to enhance conductivity and surface area [19] | Excellent electrical conductivity, high surface-to-volume ratio, functional groups for bioconjugation |
| Metal-Organic Frameworks (MOFs) | Porous nanomaterial to increase immobilization capacity and pre-concentrate analytes [19] | Extremely high surface area, tunable porosity, enhances sensor loading and sensitivity |
| Methylene Blue | Redox-active reporter label in electrochemical aptasensors [19] | Intercalates with DNA; change in signal upon aptamer conformation/ displacement indicates binding |
| Nafion | Cation-exchange polymer; used as a permselective membrane and binder for biocomposite inks [10] | Prevents fouling, stabilizes enzyme layers, binds nanomaterials to electrode surfaces |
The strategic selection of a biorecognition element is the cornerstone of developing a successful electrochemical biosensor for pesticide analysis. Enzymes offer a biologically relevant mechanism for class-specific detection, while aptamers provide a versatile and stable platform for highly specific quantification. Antibodies remain the gold standard for immunoassays requiring extreme specificity, and MIPs present a robust, cost-effective biomimetic alternative. The protocols and comparisons detailed in this application note provide a framework for researchers to make informed decisions, balancing analytical requirements with practical constraints to advance the field of food safety monitoring.
Electrochemical biosensors have emerged as powerful analytical tools for the rapid and on-site detection of pesticide residues in fruits and vegetables, aligning with the growing need for food safety monitoring [31] [32]. The performance of these biosensors is critically dependent on the electrode materials and their modification with nanomaterials to amplify the electrochemical signal. Among the various nanomaterials available, gold nanoparticles (AuNPs), graphene and its derivatives, and carbon nanotubes (CNTs) belong to an elite group of nanomaterials that significantly enhance biosensor sensitivity, stability, and overall performance [33]. This protocol details the application of these nanomaterials in constructing high-sensitivity electrochemical biosensors specifically for detecting organophosphate and carbamate pesticides in fruit samples, providing a standardized methodology for researchers and scientists in the field of food safety and analytical chemistry.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 1: Key Research Reagents and Materials for Nanomaterial-Enhanced Electrochemical Biosensors
| Reagent/Material | Function/Description | Application in Biosensor Fabrication |
|---|---|---|
| Gold Nanoparticles (AuNPs) | Colloidal solution with negative charge, large surface area, facile surface modification with thiols, low toxicity, and high biocompatibility [32]. | Provides a platform for biomolecule immobilization (antibodies, aptamers); enhances electron transfer and catalytic activity [33] [34]. |
| Graphene Oxide (GO) / Reduced GO (rGO) | Two-dimensional carbon nanomaterial with high surface area, excellent electrical conductivity, and abundant functional groups for bioconjugation [33] [35]. | Increases electroactive surface area; facilitates direct electron transfer; often used in nanocomposites to synergistically improve sensor performance [33] [34]. |
| Multi-Walled Carbon Nanotubes (MWCNTs) | Cylindrical carbon nanostructures with high electrical conductivity and mechanical strength; prone to agglomeration without functionalization [34] [35]. | Used as electrode modifiers to enhance electron transfer kinetics; often combined with other nanomaterials like rGO to form highly conductive networks [33] [34]. |
| Screen-Printed Electrodes (SPEs) | Disposable, low-cost, miniaturized electrochemical cells (working, reference, and counter electrode integrated) ideal for portable analysis [31] [36]. | Serve as the foundational substrate for nanomaterial modification and biosensor assembly; enable on-site testing with minimal sample volume [31] [37]. |
| Specific Aptamers/Antibodies | Biological recognition elements (single-stranded DNA/RNA or antibodies) with high affinity and specificity for target pesticide molecules [32]. | Immobilized on the nanomaterial-modified electrode to provide selective binding for the target analyte, forming the basis of the biosensing mechanism [31] [32]. |
| Chitosan (CS) | A biocompatible polymer with excellent film-forming ability and adhesion properties [34]. | Used as a dispersing agent for nanomaterials like MWCNTs and rGO and as a matrix for stable immobilization of biorecognition elements on the electrode surface [34]. |
Performance Comparison of Nanomaterial-Based Biosensors
The integration of nanomaterials into electrochemical biosensors has led to remarkable improvements in analytical performance for pesticide detection. The following table summarizes the reported efficacy of sensors utilizing different nanomaterial configurations.
Table 2: Analytical Performance of Nanomaterial-Enhanced Biosensors for Pesticide Detection
| Nanomaterial Configuration | Target Pesticide(s) | Detection Technique | Linear Range | Limit of Detection (LOD) | Key Advantages |
|---|---|---|---|---|---|
| Acetylcholinesterase (AChE) / c-MWCNT / Fe₃O₄-NP [32] | Malathion, Chlorpyrifos | Amperometry | Not Specified | 0.1 nM | High sensitivity; reusable >50 times; stable for 2 months. |
| Aptamer / Fe-Co MNPs / Fe-N-C Nanozyme [32] | Phorate, Profenofos | Colorimetry | Not Specified | 0.16 ng/mL (Phorate, Profenofos) | High specificity and stability; satisfactory recovery in vegetable samples. |
| MXene/Carbon Nanohorn/β-CD-MOF [32] | Carbendazim | Voltammetry | 0.003 to 10.0 μM | 1.0 nM | Excellent catalytic activity and high electronic conductivity. |
| MWCNTs-rGO-Chitosan [34] | Tau-441 Protein (Model Biomarker) | Differential Pulse Voltammetry (DPV) | 0.5 - 80 fM | 0.46 fM | Signal multi-amplification via nanomaterial synergy and AuNP labels. |
| Au–Pd / rGO / MWCNTs Nanocomposite [31] | Pesticides (General) | Voltammetry | Not Specified | Not Specified | Enhanced electrocatalytic activity and surface area from noble metals and carbon nanomaterials. |
Experimental Protocol: Fabrication of a Nanocomposite-Based Electrochemical Aptasensor
Principle
This protocol describes the construction of an electrochemical aptasensor for the detection of organophosphorus pesticides (OPPs). The sensor is based on a glassy carbon electrode (GCE) modified with a multi-walled carbon nanotube-reduced graphene oxide (MWCNTs-rGO) nanocomposite to enhance the electrode surface area and conductivity. A specific aptamer against the target OPP is immobilized on this platform. The detection mechanism relies on the change in electrochemical signal, measured via Differential Pulse Voltammetry (DPV), when the aptamer binds to its target pesticide [31] [34] [32].
Materials and Equipment
- Electrochemical Workstation: Capable of performing Cyclic Voltammetry (CV), Electrochemical Impedance Spectroscopy (EIS), and DPV.
- Electrodes: Glassy Carbon Electrode (GCE, 3 mm diameter) or Disposable Screen-Printed Carbon Electrodes (SPCEs); Ag/AgCl reference electrode; Platinum counter electrode.
- Chemicals: Multi-walled carbon nanotubes (MWCNTs), Graphene Oxide (GO) dispersion, Chitosan (CS), chloroauric acid (HAuCl₄), organophosphate pesticide (OPP) aptamer, potassium ferricyanide (K₃[Fe(CN)₆]), phosphate buffer saline (PBS, 0.1 M, pH 7.4).
- Lab Equipment: Ultrasonic bath, centrifuge, magnetic stirrer, pH meter, micropipettes.
Step-by-Step Procedure
Step 1: Synthesis of MWCNTs-rGO-Chitosan Nanocomposite
- Dispersion: Disperse 10 mg of MWCNTs and 10 mg of GO in 20 mL of deionized water separately. Sonicate both dispersions for 60 minutes until homogeneous.
- Mixing: Combine the MWCNTs and GO dispersions and continue sonication for an additional 60 minutes to form a uniform MWCNTs-GO mixture.
- Chemical Reduction: Add 100 µL of hydrazine hydrate (as a reducing agent) to the mixture and heat at 95°C for 2 hours under continuous stirring to reduce GO to rGO, resulting in an MWCNTs-rGO dispersion.
- Incorporation of Chitosan: Dissolve 0.5% (w/v) chitosan in 1% (v/v) acetic acid solution. Add 5 mL of this chitosan solution to the MWCNTs-rGO dispersion and stir for 30 minutes to form the final MWCNTs-rGO-CS nanocomposite. Store at 4°C when not in use [34].
Step 2: Electrode Pretreatment and Modification
- GCE Polishing: Polish the GCE sequentially with 1.0, 0.3, and 0.05 µm alumina slurry on a microcloth pad. Rinse thoroughly with deionized water between each polish and after the final polish.
- Electrochemical Cleaning: In a solution of 0.1 M H₂SO₄, perform cyclic voltammetry between -0.2 V and +1.0 V (vs. Ag/AgCl) at a scan rate of 100 mV/s until a stable voltammogram is obtained. Rinse the electrode with deionized water.
- Nanocomposite Coating: Deposit 8 µL of the MWCNTs-rGO-CS nanocomposite dispersion onto the clean, dry surface of the GCE. Allow it to dry under an infrared lamp or at room temperature to form a stable film. Label this as the GCE/MWCNTs-rGO-CS electrode.
Step 3: Aptamer Immobilization
- Aptamer Preparation: Dilute the synthetic OPP-specific aptamer to a concentration of 1 µM in 10 mM Tris-EDTA (TE) buffer, pH 8.0.
- Immobilization: Drop-cast 5 µL of the aptamer solution onto the surface of the GCE/MWCNTs-rGO-CS electrode.
- Incubation: Incubate the electrode in a humidified chamber at 4°C for 12-16 hours to allow for effective immobilization of the aptamer onto the nanocomposite film via electrostatic interactions and physical adsorption.
- Rinsing: Gently rinse the electrode with PBS (pH 7.4) to remove any unbound aptamer strands. The electrode is now functionalized and labeled as the Aptasensor (GCE/MWCNTs-rGO-CS/Apt).
Step 4: Electrochemical Measurement and Pesticide Detection
- Measurement Setup: Use a three-electrode system with the prepared aptasensor as the working electrode, Ag/AgCl as the reference, and a platinum wire as the counter electrode. The measurement solution is 5 mM [Fe(CN)₆]³⁻/⁴⁻ in 0.1 M PBS (pH 7.4).
- Baseline Signal Record: First, record a DPV curve for the aptasensor in the measurement solution without the target pesticide. This serves as the baseline signal (I₀). The DPV parameters are: potential range from -0.1 V to +0.5 V, pulse amplitude of 50 mV, pulse width of 50 ms.
- Sample Incubation: Incubate the aptasensor in a sample solution (e.g., extracted fruit juice) spiked with a known concentration of the target OPP for 15 minutes at room temperature.
- Post-Incubation Measurement: Rinse the electrode gently with PBS to remove non-specifically bound molecules. Record the DPV signal again (I) under the same conditions as step 2.
- Signal Analysis: The binding of the target pesticide to the immobilized aptamer causes a steric hindrance, reducing the current signal. The signal decrease (I₀ - I) is proportional to the concentration of the target pesticide in the sample.
Data Analysis
- Calibration Curve: Measure the DPV signal for a series of standard solutions with known pesticide concentrations. Plot the signal decrease (ΔI = I₀ - I) against the logarithm of the pesticide concentration.
- Quantification: Fit the data points with a linear regression model (y = a + bx). The resulting calibration curve can be used to interpolate the concentration of unknown samples based on their measured ΔI.
- Validation: Validate the sensor's accuracy by testing spiked real samples (e.g., apple or cucumber extracts) and calculating the recovery rate [32].
Workflow and Signaling Visualization
Sensor Assembly and Measurement Flow
The diagram above illustrates the sequential protocol for fabricating the aptasensor and detecting pesticides. The critical signal amplification occurs at Step 2, where the MWCNTs-rGO nanocomposite is applied. This layer enhances the electroactive surface area and facilitates electron transfer, leading to a higher initial baseline current (I₀). Upon pesticide binding (Step 5), the formation of the aptamer-pesticide complex on the nanocomposite surface acts as an insulating layer, hindering the access of the redox probe ([Fe(CN)₆]³⁻/⁴⁻) to the electrode and resulting in a measurable decrease in the DPV current (I). This change (ΔI) is the quantitative basis for detection [34] [32].
Troubleshooting and Best Practices
- Nanocomposite Film Homogeneity: To avoid uneven "coffee-ring" effects during drop-casting, ensure the nanocomposite dispersion is well-sonicated immediately before use. Optimizing the chitosan concentration can improve film stability and uniformity [35].
- Non-Specific Binding: To minimize false positives, consider blocking the modified electrode surface after aptamer immobilization with a non-interfering protein like Bovine Serum Albumin (BSA, 1% w/v) for 30 minutes.
- Reproducibility: For consistent results, standardize the drying conditions (time and temperature) for the nanomaterial modification step and use freshly prepared dispersions to prevent aggregation.
- Real Sample Analysis: For complex fruit and vegetable matrices, a simple sample pretreatment such as filtration or dilution of the juice supernatant with buffer is recommended to reduce interference [31] [32].
The performance of an electrochemical biosensor is fundamentally governed by the interface between the biological recognition element and the electrode surface. Surface functionalization—the process of modifying a solid substrate with specific chemical groups or biomolecules—is therefore a critical step in biosensor development. This process enables the precise immobilization of probes, such as oligonucleotides or enzymes, ensuring optimal orientation, density, and stability for detecting target analytes. The choice of immobilization strategy is highly dependent on the electrode material, the nature of the biological probe, the sensing environment, and the required analytical performance metrics such as sensitivity, stability, and specificity [38] [39]. Within the context of detecting fruit pesticide residues, a robust and well-engineered sensor surface is paramount for achieving reliable measurements in complex sample matrices.
This protocol details the primary workflows for functionalizing various electrode materials and immobilizing biological probes, with a specific focus on applications in food safety and pesticide residue analysis. The methods are designed to provide researchers with a comprehensive toolkit for constructing high-performance electrochemical biosensors.
Core Principles and Immobilization Methodologies
The immobilization of biological probes onto transducer surfaces can be achieved through physical adsorption, covalent bonding, or bio-affinity interactions. Each method offers distinct advantages and limitations, summarized in Table 1 below.
Table 1: Comparison of Probe Immobilization Techniques for Electrochemical Biosensors
| Immobilization Technique | Mechanism of Interaction | Key Advantages | Key Limitations | Applicable Electrode Materials |
|---|---|---|---|---|
| Physical Adsorption | Hydrophobic interactions, ionic bonding, van der Waals forces [40]. | Simple procedure, no chemical modifiers required [40]. | Weak binding, prone to probe leakage and random orientation [40]. | Carbon, Graphene, Polymers |
| Covalent Binding | Formation of stable covalent bonds (e.g., Au-Thiol, amine-carboxyl) [38] [39]. | Stable, robust layers; controlled probe density and orientation [38]. | Requires chemical modification of probe and/or surface; multi-step process [41]. | Gold, Carbon, Functionalized Polymers |
| Avidin-Biotin Affinity | High-affinity non-covalent interaction between (strept)avidin and biotin [38] [40]. | Very strong binding; versatile; suitable for various biomolecules [39]. | Requires biotinylated probes; streptavidin tetramer can cause steric hindrance [39]. | Any surface where avidin/streptavidin can be adsorbed or covalently linked |
| Self-Assembled Monolayers (SAMs) | Spontaneous organization of molecules (e.g., thiols on gold) into ordered layers [38] [39]. | Highly ordered and reproducible surfaces; enables surface passivation [39]. | Can exhibit baseline signal drift; stability can be time-dependent [39]. | Gold, Platinum, other metals |
Material-Specific Functionalization Protocols
The chemical nature of the electrode material dictates the most effective functionalization strategy. The following protocols cover the most common materials used in electrochemical biosensors.
Gold Electrodes: Thiol-Based Self-Assembly
Gold is one of the most extensively studied electrode materials due to its excellent conductivity and the well-established chemistry of gold-thiol self-assembled monolayers (SAMs) [38] [39].
Protocol: Thiolated DNA Probe Immobilization on Gold
- Reagents: Thiol-modified DNA/aptamer probe, 6-Mercapto-1-hexanol (MCH), Absolute ethanol, Phosphate Buffered Saline (PBS, pH 7.4).
- Equipment: Gold disk working electrode (e.g., 2 mm diameter), Potentiostat, Microcentrifuge tubes.
- Electrode Pretreatment: Clean the gold electrode mechanically (using alumina slurry, 0.05 µm) and electrochemically (via cyclic voltammetry in 0.5 M H₂SO₄ from -0.2 to +1.5 V until stable CV curves are obtained) to ensure a pristine surface [40]. Rinse thoroughly with deionized water and dry under a stream of nitrogen.
- Probe Immobilization: Prepare a 1 µM solution of the thiolated DNA probe in PBS. Deposit a 10 µL droplet of this solution onto the cleaned gold electrode surface. Incubate in a humidified chamber for a minimum of 1 hour at room temperature to allow for the formation of a covalent Au-S bond.
- Surface Backfilling: After incubation, rinse the electrode gently with PBS to remove physisorbed probes. Then, immerse the electrode in a 1 mM solution of MCH in ethanol for 30-60 minutes. This critical step displaces nonspecifically adsorbed probes and creates a well-ordered, passivated monolayer that minimizes non-specific binding and aligns the DNA probes upright for better target accessibility [38] [40].
- Rinsing and Storage: Rinse the functionalized electrode thoroughly with PBS and deionized water. The sensor is now ready for use and should be stored in PBS at 4°C if not used immediately.
Carbon-Based Electrodes: Covalent and Affinity Strategies
Carbon materials (glassy carbon, screen-printed carbon) are widely used due to their broad potential window, low cost, and biocompatibility. However, their functionalization requires different approaches [38].
Protocol A: Carbodiimide Crosslinking for Amine-Terminated Probes
- Reagents: 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS), Amine-modified DNA probe or antibody, Acetate buffer (0.1 M, pH 5.0).
- Equipment: Carbon working electrode, Potentiostat.
- Electrode Activation: Polish the carbon electrode with alumina slurry and rinse. To create carboxyl groups on the surface, perform anodic polarization (e.g., +1.5 V vs. Ag/AgCl for 5 min in 0.1 M NaOH) or use ultraviolet-ozone treatment [39].
- NHS/EDC Activation: Prepare a fresh mixture of 400 mM EDC and 100 mM NHS in acetate buffer. Apply this solution to the activated carbon electrode and incubate for 30-60 minutes. This step converts the surface carboxyl groups into amine-reactive NHS esters.
- Probe Conjugation: Rinse the electrode with acetate buffer to remove excess EDC/NHS. Immediately apply a solution of the amine-terminated probe (e.g., 1-10 µM in PBS) and incubate for 2 hours. The amine groups on the probe will form stable amide bonds with the activated ester on the electrode surface.
- Quenching and Rinsing: Quench any remaining active esters by incubating with 1 M ethanolamine (pH 8.5) for 15 minutes. Rinse with PBS before use.
Protocol B: Streptavidin-Biotin Affinity Immobilization
- Streptavidin Coating: Physically adsorb streptavidin onto the carbon electrode by applying a 0.1 mg/mL solution in PBS for 1 hour [39]. Alternatively, streptavidin can be covalently linked to a pre-activated carbon surface (as in Protocol A).
- Probe Immobilization: Rinse the streptavidin-coated electrode and incubate with a biotinylated DNA probe or antibody (e.g., 1 µM in PBS) for 30-45 minutes. The strong affinity between streptavidin and biotin (KD ≈ 10⁻¹⁵ M) will immobilize the probe.
Advanced Functionalization: 3D Nanostructures for Enhanced Sensitivity
Incorporating three-dimensional (3D) nanostructures on the electrode surface dramatically increases the surface area available for probe immobilization, leading to higher probe loading and enhanced signal amplification [42].
Protocol: Electrodeposition of Gold Nanoparticles (AuNPs) for 3D Sensing
- Reagents: Chloroauric acid (HAuCl₄), Potassium chloride (KCl).
- Equipment: Potentiostat, Conventional three-electrode system.
- Solution Preparation: Prepare an electrodeposition solution containing 1 mM HAuCl₄ and 0.1 M KCl.
- Electrodeposition: Immerse the clean working electrode (e.g., glassy carbon or gold) into the solution. Apply a constant potential of -0.4 V (vs. Ag/AgCl reference) for 30-120 seconds. The reduction of Au³⁺ to Au⁰ will result in the formation of a nanostructured AuNP layer on the electrode surface [42].
- Characterization: The deposited AuNP layer can be characterized by scanning electron microscopy (SEM) and cyclic voltammetry.
- Probe Immobilization: The AuNP-modified electrode can now be functionalized with thiolated probes using the standard gold-thiol protocol described in Section 2.2.1. The 3D architecture provides a significantly higher density of immobilization sites.
Application in Pesticide Residue Biosensing
The functionalization workflows described above are directly applicable to the development of biosensors for fruit pesticide residues. A prominent example is the construction of an enzymatic biosensor for organophosphorus pesticides (OPs), which operates on an inhibition principle [10].
Experimental Protocol: On-Glove Enzymatic Biosensor for Pesticide Detection
- Objective: To detect organophosphorus pesticides (e.g., dichlorvos) directly on fruit peels via enzyme inhibition [10].
- Biosensor Design: An electrochemical biosensor is fabricated on a screen-printed electrode (SPE) integrated onto a finger of a glove.
- Functionalization Workflow:
- Electrode Modification: The carbon-based SPE is first modified with a composite of Prussian blue (electron mediator) and carbon black (nanostructured conductor) to enhance the electrochemical signal.
- Enzyme Immobilization: The enzyme butyrylcholinesterase (BChE) is physically adsorbed onto the modified electrode surface. The enzyme serves as the biological recognition element.
- Detection Principle: The active BChE enzyme hydrolyzes its substrate (e.g., butyrylthiocholine), producing an electroactive product that is detected amperometrically. In the presence of an OP pesticide, the enzyme is inhibited, leading to a measurable decrease in the electrochemical signal. The degree of inhibition is proportional to the pesticide concentration [10].
- On-Site Application: The end-user simply scrubs the finger containing the functionalized electrode against the fruit peel, then adds a drop of substrate solution to perform the electrochemical reading, enabling decentralized analysis directly in the field.
Performance Evaluation and Metrics
The analytical performance of a functionalized biosensor is validated by assessing key figures of merit. Table 2 summarizes these parameters and the common methods for their evaluation.
Table 2: Key Analytical Figures of Merit for Biosensor Validation [43]
| Figure of Merit | Definition | Evaluation Method |
|---|---|---|
| Sensitivity | The slope of the analytical calibration curve (signal vs. concentration). | Measured from the linear range of the calibration plot. High sensitivity is indicated by a large change in signal for a small change in concentration. |
| Limit of Detection (LOD) | The lowest concentration of analyte that can be reliably distinguished from zero. | Typically calculated as 3× (standard deviation of the blank) / sensitivity. |
| Selectivity | The ability to distinguish the target analyte from potential interferents. | Measuring the sensor response in the presence of structurally similar compounds or common matrix components. |
| Repeatability | Closeness of agreement between successive measurements under identical conditions. | Expressed as the relative standard deviation (RSD%) of multiple measurements of the same sample. |
| Reproducibility | Closeness of agreement between measurements under changed conditions (e.g., different operators, days). | Expressed as the RSD% of measurements performed in the changed conditions. |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Probe Immobilization and Surface Functionalization
| Reagent / Material | Function in Fabrication Workflow | Common Examples / Notes |
|---|---|---|
| Thiol-/Amino-modified Oligonucleotides | To provide a terminal chemical handle for covalent immobilization on specific surfaces. | Thiol for gold surfaces [38]; Amine for EDC/NHS chemistry on carboxylated surfaces [39]. |
| EDC and NHS | Carbodiimide crosslinkers for activating carboxyl groups to form amide bonds with amines. | Must be prepared fresh; EDC is unstable in aqueous solution [39] [41]. |
| Mercaptoalkanol (e.g., MCH) | Used as a backfilling agent in SAMs on gold to passivate the surface and orient probes. | Reduces non-specific adsorption and prevents probe lying down [38] [40]. |
| Streptavidin/Avidin | Protein used as a bridge for immobilizing biotinylated probes via high-affinity binding. | Provides a versatile and strong immobilization platform [38] [39]. |
| Prussian Blue & Carbon Black | Redox mediator and conductive nanomaterial, respectively, to enhance electrochemical signal. | Used in enzymatic biosensors for signal amplification, e.g., in pesticide sensors [10]. |
| Gold Nanoparticles (AuNPs) | Nanomaterials to create 3D electrode surfaces, increasing probe loading and sensitivity. | Can be electrodeposited or drop-casted [42]. |
Workflow Visualization
The following diagram summarizes the decision-making workflow for selecting an appropriate surface functionalization strategy based on the electrode material and the desired application.
Biosensor Fabrication Workflow Selection
The accurate measurement of pesticide residues in fruit matrices is a critical component of food safety monitoring. This protocol details a stepwise analytical procedure for sample preparation, from fruit collection to final measurement, specifically framed within research on electrochemical biosensors. Traditional methods often rely on complex, time-consuming laboratory techniques, but the emergence of novel green extraction methods and portable detection technologies, such as on-glove biosensors, offers new possibilities for rapid, on-site analysis [44] [10]. This document provides detailed methodologies to support researchers and scientists in developing robust and efficient analytical workflows.
Sample preparation is the most critical stage in the analytical workflow, directly impacting the accuracy, sensitivity, and precision of the subsequent detection method [18]. The primary goal is to isolate the target analytes (pesticides) from the complex fruit matrix while minimizing co-extractives that can interfere with the analysis.
The table below summarizes the fundamental principles, advantages, and limitations of several established and innovative sample preparation techniques.
Table 1: Comparison of Sample Preparation Techniques for Pesticide Residue Analysis
| Technique | Fundamental Principle | Key Advantages | Potential Limitations |
|---|---|---|---|
| Pressurized Liquid Extraction (PLE) | Uses liquid solvents at elevated temperatures and pressures [44]. | High extraction efficiency, faster extraction times, reduced solvent consumption [44]. | Requires specialized equipment, potential for thermal degradation of some analytes. |
| Supercritical Fluid Extraction (SFE) | Utilizes supercritical fluids (e.g., CO₂) as the extraction solvent [44]. | Eliminates organic solvents, high selectivity, easily tunable parameters [44]. | High initial equipment cost, can be less effective for very polar pesticides. |
| Gas-Expanded Liquid Extraction (GXL) | Involves expanding a liquid solvent with a compressed gas (e.g., CO₂) [44]. | Combines advantages of liquid and supercritical solvents, improved mass transfer [44]. | Relatively new technique, process optimization can be complex. |
| Microextraction Techniques | Miniaturized extraction using a very small volume of solvent [18]. | Minimal solvent consumption, simplicity, can be integrated into field-deployable devices [18]. | May require careful optimization for different fruit matrices. |
Detailed Protocol: QuEChERS Method for Fruit Matrices
The QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) method is a widely used sample preparation technique for multiresidue pesticide analysis in fruits and vegetables. The following is a detailed protocol.
Materials and Equipment
- Homogenizer (e.g., blender or food processor)
- Centrifuge tubes (50 mL, with screw caps)
- Analytical balance
- Centrifuge
- Vortex mixer
- Volumetric pipettes and dispensers
- Acetonitrile (HPLC grade)
- Acetic Acid (or formic acid) for buffering
- Anhydrous Magnesium Sulfate (MgSO₄)
- Sodium Chloride (NaCl)
- Dispersive SPE (d-SPE) kits (e.g., containing PSA, C18, and MgSO₄)
Step-by-Step Procedure
- Sample Homogenization: Weigh 15 g of a representative, homogenized fruit sample into a 50 mL centrifuge tube.
- Solvent Addition: Add 15 mL of acetonitrile (with 1% acetic acid) to the tube.
- Shaking: Cap the tube and shake vigorously for 1 minute to ensure the solvent thoroughly interacts with the sample.
- Salt Addition: Add a pre-packaged salt mixture (typically 6 g of MgSO₄ and 1.5 g of NaCl) to induce liquid-liquid partitioning. Immediately shake the tube for another minute to prevent salt clumping.
- Centrifugation: Centrifuge the tubes at >4000 RPM for 5 minutes to separate the organic (acetonitrile) layer from the fruit solids and water.
- Clean-up (d-SPE): Transfer an aliquot (e.g., 1 mL) of the upper acetonitrile layer to a d-SPE tube containing 150 mg MgSO₄ and 25 mg Primary Secondary Amine (PSA) sorbent. PSA helps remove fatty acids and sugars, while MgSO₄ removes residual water.
- Vortex and Centrifuge: Vortex the d-SPE tube for 30 seconds and centrifuge. The supernatant is now ready for analysis.
Detailed Protocol: On-Site Sampling for Portable Biosensors
For direct analysis using portable electrochemical biosensors, the sample preparation is significantly simplified, focusing on transferring the analyte from the fruit surface to the sensor.
Materials and Equipment
- On-glove electrochemical biosensor (e.g., integrating a screen-printed electrode modified with Prussian blue, Carbon black, and butyrylcholinesterase enzyme) [10].
- Portable potentiostat for electrochemical readings.
- Blunt-ended swabs or wipes (if not integrated into the sensor).
Step-by-Step Procedure
- Sensor Preparation: If using a disposable sensor, unpack and initialize it according to the manufacturer's instructions. For a reusable on-glove system, ensure the electrode surface is clean and functional [10].
- Direct Sampling: The end-user simply scrubs the surface of the fruit (e.g., apple, orange) with the integrated sampling strip on the glove [10]. The pesticide residues are transferred directly from the fruit peel to the biosensor surface.
- Measurement: Perform the electrochemical reading immediately after sampling using the portable potentiostat. The detection is often based on the inhibition of the enzyme (e.g., butyrylcholinesterase) in the presence of the pesticide (e.g., dichlorvos) [10].
Workflow Visualization
The following diagram illustrates the complete analytical procedure, from sample collection to final measurement, highlighting the two main pathways (conventional lab-based vs. on-site analysis).
Workflow for Fruit Pesticide Analysis
The Scientist's Toolkit: Research Reagent Solutions
The following table details key reagents and materials essential for the experiments described in these protocols.
Table 2: Essential Research Reagents and Materials for Pesticide Residue Analysis
| Item | Function / Role in the Protocol |
|---|---|
| Butyrylcholinesterase Enzyme | Biological recognition element in inhibition-based electrochemical biosensors; its activity is inhibited by organophosphorus pesticides, enabling detection [10]. |
| Prussian Blue & Carbon Black | Redox mediator and nanostructured material used to modify electrode surfaces; they enhance the electron transfer rate and improve the sensitivity of the biosensor [10]. |
| Acetonitrile | Common extraction solvent used in methods like QuEChERS due to its ability to precipitate proteins and extract a wide range of pesticides from aqueous fruit matrices. |
| Primary Secondary Amine (PSA) | A sorbent used in the clean-up step (d-SPE) to remove interfering compounds such as fatty acids and sugars from the fruit extract. |
| Anhydrous Magnesium Sulfate (MgSO₄) | Used as a drying salt to remove residual water from the organic extract during the partitioning and clean-up steps, improving recovery and stability. |
| Deep Eutectic Solvents (DES) | Novel, green solvents with low toxicity and high biodegradability; emerging as sustainable alternatives to conventional organic solvents for extraction [44]. |
| Supercritical CO₂ | The extraction fluid in Supercritical Fluid Extraction (SFE); it is non-toxic, non-flammable, and provides high penetration into the sample matrix [44]. |
Electrochemical biosensors incorporating screen-printed electrodes (SPEs) represent a transformative technology for decentralized food safety analysis, aligning with the principles of precision agriculture. These sensors address critical limitations of conventional chromatographic methods, which are laboratory-bound, time-consuming, and require specialized personnel [45]. The integration of SPEs onto wearable platforms, such as gloves, marks a significant advancement towards real-time, on-site monitoring of pesticide residues, enabling proactive risk assessment directly in the field or at points of inspection [10] [46]. This document details the application and protocol for an innovative on-glove biosensor, providing a practical framework for researchers developing electrochemical biosensor protocols for fruit pesticide residue analysis.
Detailed Case Study: On-Glove Biosensor for Fruit Analysis
Sensor Concept and Workflow
This case study focuses on an enzymatic inhibition biosensor engineered onto a glove for the quantification of organophosphorus (OP) pesticides directly on fruit peels [10] [47]. The detection principle relies on the inhibition of the enzyme butyrylcholinesterase (BChE) in the presence of OP pesticides like dichlorvos. The bio-hybrid sensing probe integrates Prussian blue, Carbon black, and the BChE enzyme on a screen-printed electrode platform attached to a glove fingertip [10]. The operational workflow is visually summarized in the diagram below.
Performance and Comparative Analysis
The on-glove biosensor demonstrated high sensitivity and practical applicability. Its performance against other SPE-based sensor configurations is detailed in the following table.
Table 1: Performance Metrics of SPE-Based Sensors for Pesticide Detection
| Sensor Configuration | Target Pesticide (Class) | Detection Principle | Linear Range | Limit of Detection (LOD) | Tested Matrices |
|---|---|---|---|---|---|
| On-Glove Biosensor [10] | Dichlorvos (Organophosphorus) | Enzymatic Inhibition (BChE) | Nanomolar range | Nanomolar (high ppt), lower than EU MRL | Apple and orange peels |
| Multi-Analyte Glove Sensor [48] | Carbamates, Phenylamides, Bipyridinium, Organophosphates | Not Specified | Not Specified | Not Specified | Food products, fruit juice |
| Aptamer-Based Sensor [32] | Carbendazim | MXene/Carbon Nanohorn/β-CD-MOF | 0.003 to 10.0 µM | 1.0 nM | Not Specified |
| Fe3O4-NP/AChE Biosensor [32] | Malathion, Chlorpyrifos (Organophosphorus) | Enzymatic Inhibition (AChE) | Not Specified | 0.1 nM | Not Specified |
Abbreviations: MRL (Maximum Residue Limit), EU (European Union), BChE (Butyrylcholinesterase), AChE (Acetylcholinesterase), β-CD-MOF (β-Cyclodextrin-Metal-Organic Framework), NP (Nanoparticle).
Key outcomes from the on-glove sensor application include:
- High Sensitivity: The sensor achieved a detection limit for dichlorvos in the nanomolar range (high parts-per-trillion), which is lower than the maximum residue limits established by European Union regulations [10].
- Excellent Repeatability: The system demonstrated satisfactory analytical repeatability with a relative standard deviation (RSD) lower than 10% [10] [47].
- Matrix Application: The methodology was successfully validated for the direct detection of pesticides on the complex surfaces of apples and oranges without extensive sample pre-treatment [10].
Experimental Protocol: On-Glove Detection of Organophosphorus Pesticides
Research Reagent Solutions and Materials
The following table lists the essential materials and reagents required to replicate the on-glove biosensor experiment.
Table 2: Essential Research Reagents and Materials for On-Glove Biosensor Fabrication
| Item Name | Specification / Function | Application in Protocol |
|---|---|---|
| Screen-Printed Electrode (SPE) | Ceramic/plastic substrate with carbon, silver, or gold ink [49] [45] | Serves as the disposable, miniaturized electrochemical transducer platform. |
| Butyrylcholinesterase (BChE) | Enzyme; inhibition by OPs is the basis for detection [10] [45] | Biological recognition element immobilized on the working electrode. |
| Prussian Blue | Mediator; electrocatalyst for low-potential detection [10] | Enhances electron transfer and amplifies the electrochemical signal. |
| Carbon Black | Nanomaterial; high surface area and conductivity [10] | Increases the active surface area of the electrode, improving sensitivity. |
| Dichlorvos Standard | Organophosphorus pesticide; model analyte [10] | Used for sensor calibration and performance evaluation. |
| Portable Potentiostat | Compact electronic instrument for electrochemical measurements [45] | Provides the potential and measures the current for analysis in the field. |
| Nitrile Glove | Substrate for sensor integration; less porous than latex [48] | Wearable platform that holds the SPE sensors on the fingertips. |
Step-by-Step Experimental Procedure
Part A: Biosensor Fabrication and Glove Integration
- SPE Modification: Prepare the bio-hybrid ink by thoroughly mixing Prussian blue, Carbon black, and the butyrylcholinesterase (BChE) enzyme in a suitable buffer solution [10].
- Immobilization: Drop-cast a precise volume of the prepared bio-hybrid ink onto the working electrode surface of the screen-printed electrode. Allow it to dry under controlled conditions (e.g., at room temperature or 4°C) to form a stable film [10] [45].
- Glove Assembly: Integrate the modified SPE onto the index finger of a nitrile glove using a biocompatible adhesive, ensuring that the electrode contacts are accessible for connection to the potentiostat [10] [48].
Part B: Electrochemical Measurement and Pesticide Detection
- System Setup: Connect the fingers of the equipped glove, bearing the integrated SPE, to a portable potentiostat.
- Sample Collection: The end-user simply scrubs the surface of the target fruit (e.g., apple, orange) with the sensor-modified fingertip. This action transfers the pesticide residues from the fruit peel to the sensor surface [10].
- Electrochemical Analysis: Initiate the chronoamperometric measurement using the portable potentiostat. The applied potential should be optimized for the Prussian blue-mediated system.
- Signal Interpretation: Measure the steady-state current. The presence of an organophosphorus pesticide will inhibit the BChE enzyme, leading to a measurable decrease in the catalytic current. The degree of current suppression is proportional to the pesticide concentration [10] [45].
The relationship between enzyme activity, inhibition, and the measured signal is illustrated below.
The integration of screen-printed electrodes into wearable formats like the described on-glove biosensor demonstrates a powerful application of electrochemical biosensing for decentralized food safety analysis. This protocol provides a reproducible methodology for detecting organophosphorus pesticides directly on fruit peels, characterized by its simplicity for the end-user, high sensitivity, and rapid results. This technology serves as a robust model for the development of future on-site diagnostic tools in precision agriculture and food safety monitoring, paving the way for broader applications in environmental and health diagnostics [10] [46].
Maximizing Performance: Strategies for Sensitivity, Selectivity, and Real-World Reliability
Overcoming Matrix Effects in Complex Fruit Samples
Electrochemical biosensors represent a transformative technology for rapid, on-site detection of pesticide residues in agricultural products. However, their application to complex fruit matrices presents significant challenges due to matrix effects—interferences from fruit components such as organic acids, sugars, phenolic compounds, and pigments that can alter sensor response, reduce sensitivity, and generate false positives or negatives [50] [14]. These effects primarily arise from non-specific binding of interferents to sensor surfaces, fouling of electrode interfaces, and competitive binding that masks target analyte detection [37] [51].
This protocol details specialized methodologies to overcome these limitations, leveraging innovations in sample preparation, sensor design, and interface engineering. The approaches described herein enable reliable quantification of organophosphorus pesticides and other contaminants directly on fruit peels and in fruit extracts, facilitating precise monitoring aligned with global food safety regulations [10] [50].
Experimental Protocols
On-Glove Biosensor Application for Direct Fruit Peel Analysis
This protocol, adapted from Talanta (2025), describes a minimally-invasive approach for detecting organophosphorus pesticides directly on fruit surfaces using biosensors integrated onto gloves [10].
Materials and Equipment
- Nitrile gloves (chemical-resistant)
- Screen-printed carbon electrodes (SPCEs) fabricated with Prussian blue, carbon black, and butyrylcholinesterase enzyme [10]
- Portable potentiostat for electrochemical measurements
- Dichlorvos standards (or other target organophosphorus pesticides)
- Phosphate buffer saline (PBS) (0.1 M, pH 7.4)
- Fruit samples (apples, oranges, etc.)
Procedure
- Sensor Integration: Affix the modified SPCEs to the index finger of nitrile gloves using biocompatible adhesive, ensuring the electrode surface remains fully exposed for contact with fruit peels.
- Sampling: Gently scrub the fruit peel surface (approximately 4 cm² area) for 30 seconds using the sensor-integrated glove finger, applying minimal pressure to transfer pesticide residues to the sensor interface.
- Electrochemical Measurement: Following sampling, connect the glove-integrated sensor to a portable potentiostat and perform chronoamperometric measurements at an applied potential of +0.7 V vs. pseudo-Ag/AgCl in PBS.
- Inhibition Calculation: Quantify pesticide concentration based on the inhibition of butyrylcholinesterase enzyme activity, calculated as:
% Inhibition = [(Icontrol - Isample)/Icontrol] × 100, where Icontrol represents the current response in pesticide-free conditions. - Calibration: Generate a calibration curve using standard dichlorvos solutions with concentrations ranging from 10⁻⁹ M to 10⁻⁵ M directly applied to the sensor interface.
Table 1: Performance Characteristics of On-Glove Biosensor for Dichlorvos Detection
| Parameter | Value | Conditions |
|---|---|---|
| Detection Limit | Nanomolar range (high ppt) | Apple and orange peels |
| Repeatability | <10% RSD | Multiple measurements (n=5) |
| Analysis Time | <5 minutes | Per sample |
| Linear Range | 10⁻⁹ M to 10⁻⁵ M | Dichlorvos standards |
Sample Pre-treatment Protocol for Complex Fruit Matrices
For fruit varieties with particularly high sugar content or pigment concentration, this pre-treatment protocol effectively minimizes matrix effects before electrochemical analysis [50] [14].
Materials
- Acetonitrile (HPLC grade)
- Primary secondary amine (PSA) sorbent
- Magnesium sulfate (anhydrous)
- Citrate buffer salts
- Centrifuge tubes (50 mL)
- Centrifuge (capable of 5000 × g)
- Syringe filters (0.22 μm nylon)
Procedure
- Homogenization: Homogenize 10 g of fruit tissue with 10 mL acetonitrile containing 1% acetic acid for 3 minutes at high speed.
- Extraction: Add 1.5 g MgSO₄ and 0.5 g NaCl to the homogenate, vortex vigorously for 1 minute, then centrifuge at 5000 × g for 5 minutes.
- Clean-up: Transfer 1 mL supernatant to a 2 mL microcentrifuge tube containing 50 mg PSA and 150 mg MgSO₄. Vortex for 30 seconds and centrifuge at 10,000 × g for 2 minutes.
- Dilution: Dilute the purified extract 1:1 with 0.1 M PBS (pH 7.4) to ensure compatibility with electrochemical biosensors.
- Analysis: Apply 50 μL treated sample to the electrochemical biosensor for pesticide quantification.
Table 2: Effectiveness of Sample Pre-treatment Protocols for Different Fruit Types
| Fruit Matrix | Major Interferents | Recommended Clean-up | Matrix Effect Reduction |
|---|---|---|---|
| Apple | Malic acid, polyphenols | PSA + MgSO₄ | >85% |
| Orange | Citric acid, ascorbic acid, pigments | PSA + C18 + MgSO₄ | >80% |
| Grape | Tartaric acid, sugars, anthocyanins | Z-Sep + MgSO₄ | >75% |
| Banana | Polyphenols, dopamine | PSA + MgSO₄ | >70% |
Signal Amplification and Interference Mitigation Strategies
Nanomaterial-Enhanced Sensor Interfaces
Incorporating nanomaterials into sensor design significantly reduces matrix effects by providing tailored surface properties that preferentially bind target analytes over interferents [50].
Key Materials and Functions:
- Prussian blue nanoparticles: Serve as electron transfer mediators and electrocatalysts, lowering operating potentials to reduce interferent oxidation [10].
- Carbon black: Increases electrode surface area and enhances electron transfer kinetics, improving sensitivity in complex matrices [10].
- Gold nanoparticles: Facilitate efficient antibody immobilization while repelling non-specific binding through surface charge modulation [51] [50].
- Molecularly imprinted polymers (MIPs): Create synthetic recognition sites with high selectivity for target pesticides, significantly reducing interference from fruit matrix components [51] [50].
Advanced Electrochemical Techniques
Employing pulsed voltammetric techniques rather than constant potential methods significantly reduces fouling and minimizes charging currents that mask analytical signals in complex fruit matrices [51] [50].
Optimal Techniques:
- Differential Pulse Voltammetry (DPV): Applying potential pulses with incremental voltage steps minimizes non-faradaic currents, enhancing sensitivity for pesticide detection in fruit extracts.
- Square Wave Voltammetry (SWV): Using high-frequency square waves effectively discriminates against capacitive currents, improving signal-to-noise ratios in pigmented fruit samples.
- Electrochemical Impedance Spectroscopy (EIS): Monitoring impedance changes at electrode interfaces enables detection even in heavily fouling conditions, though with potentially reduced sensitivity.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for Electrochemical Biosensing in Fruit Matrices
| Item | Function | Application Notes |
|---|---|---|
| Butyrylcholinesterase Enzyme | Biological recognition element for OPs | Inhibition-based detection; immobilization stability critical [10] |
| Screen-Printed Carbon Electrodes (SPCEs) | Disposable transducer platform | Cost-effective; customizable surface chemistry [10] [51] |
| Primary Secondary Amine (PSA) Sorbent | Matrix clean-up | Removes sugars, organic acids, and phenolic compounds [14] |
| Prussian Blue Nanoparticles | Electron transfer mediator | Lowers operating potential, reducing interferent oxidation [10] |
| Gold Nanoparticles | Signal amplification | Enhantibody immobilization; increases electroactive surface area [51] [50] |
| Molecularly Imprinted Polymers (MIPs) | Synthetic recognition elements | High chemical stability in complex fruit matrices [51] |
| Phosphate Buffer Saline (PBS) | Electrochemical buffer | Maintains pH and ionic strength during measurement [10] |
Data Interpretation and Validation
Quantification and Matrix Effect Calculation
Accurate quantification requires comparing sensor responses between standard solutions and fruit matrix samples to calculate and correct for matrix effects [37] [50].
Matrix Effect Calculation:
Where:
- Slope_matrix = Calibration curve slope in fruit extract
- Slope_standard = Calibration curve slope in pure buffer
Interpretation:
- |Matrix Effect| < 20%: Minimal suppression/enhancement
- |Matrix Effect| 20-50%: Moderate matrix effect requiring calibration with matrix-matched standards
- |Matrix Effect| > 50%: Strong matrix effect necessitating additional clean-up procedures
Method Validation Parameters
For reliable results, validate methods using these key parameters:
- Accuracy: 70-120% recovery for spiked samples
- Precision: <15% relative standard deviation (RSD)
- Limit of Detection (LOD): Typically nanomolar range for organophosphorus pesticides in fruit matrices [10]
- Selectivity: <10% cross-reactivity with similar compounds
The protocols and methodologies described herein provide researchers with robust strategies to overcome matrix effects when applying electrochemical biosensors to complex fruit samples. By implementing these specialized approaches, scientists can achieve reliable, sensitive detection of pesticide residues that meets regulatory standards while leveraging the portability and rapid analysis capabilities of electrochemical biosensing platforms.
The accurate detection of low-abundance analytes, such as pesticide residues in fruit, is a significant challenge in analytical science. Signal amplification strategies are crucial for enhancing the sensitivity and reliability of electrochemical biosensors. Nanozymes, redox cycling, and catalytic cascades represent three powerful approaches that amplify detectable signals, enabling the detection of target molecules at concentrations far below the maximum residue limits (MRLs) set by food safety authorities [52] [14]. These strategies transform the complex chemical analysis of pesticides into simpler biochemical readouts that can be deployed for rapid, on-site screening, offering a viable alternative to traditional laboratory-based methods like gas chromatography-mass spectrometry (GC-MS) and high-performance liquid chromatography (HPLC) [52] [53].
This protocol details the application of these signal amplification strategies within the context of developing electrochemical biosensors for detecting carbamate and organophosphorus pesticides in fruit samples. The integration of these methods creates a robust, anti-interference detection platform suitable for field use by researchers and food safety professionals.
Core Amplification Strategies and Mechanisms
Nanozymes: Synthetic Enzyme Mimetics
Nanozymes are nanomaterial-based catalysts that mimic the catalytic activities of natural enzymes while offering superior stability, cost-effectiveness, and versatility [54]. They overcome key limitations of natural enzymes, such as sensitivity to harsh environmental conditions (e.g., high temperature, extreme pH), difficult preparation, and special storage requirements [54]. Common nanozymes exhibit peroxidase (POD)-like, oxidase (OXD)-like, or catalase (CAT)-like activities.
Organic-dominated nanozymes are particularly advantageous for agricultural and food-sensing applications. They are synthesized from organic components like polymers, peptides, or supramolecular assemblies, which play the dominant structural and functional role in catalysis [54]. Compared to their inorganic counterparts, they offer enhanced biocompatibility, lower toxicity, and a more streamlined fabrication process that is suitable for mass production [54].
Table 1: Comparison of Selected Nanozymes for Pesticide Detection
| Nanozyme Type | Synthesis Method | Enzyme-like Activity | Detection Target | Key Advantage |
|---|---|---|---|---|
| BSA-Protected Gold Nanozymes (AuNEs) [52] | One-pot synthesis using BSA as reducing/stabilizing agent | POD-like / Intrinsic Fluorescence | Carbamate Pesticides | Dual colorimetric/fluorometric output |
| Fe₇S₈ Nanoflakes (NFs) [53] | Facile hydrothermal method | POD-like | Organophosphorus Pesticides | High stability, large surface area |
| Core-Shell Pd@Pt Nanoparticles [55] | Sonication-assisted chemical reduction | POD-like | Organophosphorus Pesticides | High synergistic catalytic activity |
| PEI-DHB Polymer Nanozyme [54] | Mixing precursors at room temperature | POD-like | General biosensing | Metal-free, simple green synthesis |
Redox Cycling: Electrochemical Amplification
Redox cycling is an electrochemical phenomenon that provides inherent signal amplification, making it highly attractive for biosensing applications where low detection limits are critical [56] [57]. In a typical configuration, an electrochemical cell features two working electrodes in close proximity—a generator and a collector—biased at different potentials.
The mechanism involves the repeated reduction and oxidation of a single electroactive molecule as it shuttles between the two electrodes. One electrode (the generator) oxidizes the molecule (R → O + e⁻), and the product (O) diffuses to the second electrode (the collector), where it is reduced back to its original state (O + e⁻ → R). This "recycling" of the analyte allows each molecule to transfer multiple electrons per unit time, resulting in a significantly higher measured current compared to a conventional single-working-electrode system [56] [57]. This configuration can yield a signal an order of magnitude larger than conventional transducers and produces a stable, non-decaying current, which improves the signal-to-noise ratio [57].
Catalytic Cascades: Multi-Enzyme Amplification
Catalytic cascade reactions integrate multiple enzymatic steps in a sequential manner, where the product of one reaction serves as the substrate for the next. This strategy effectively amplifies the initial signal by transforming a single recognition event into a large, measurable output. For pesticide detection, the Acetylcholinesterase (AChE) and Choline Oxidase (CHO) cascade is most commonly employed [52] [53] [55].
In this cascade:
- AChE hydrolyzes the neurotransmitter Acetylcholine (ACh) to produce Choline.
- CHO then oxidizes Choline, generating Betaine and Hydrogen Peroxide (H₂O₂) as a by-product.
- The concentration of H₂O₂, which is directly proportional to the activity of the AChE/CHO cascade, is then measured.
Pesticides like carbamates and organophosphates act as AChE inhibitors. Their presence reduces the activity of AChE, leading to a decrease in the amount of H₂O₂ generated. This inhibition is the measurable signal that correlates with pesticide concentration [52] [53].
Integrated Experimental Protocols
This section provides a detailed methodology for implementing a self-calibrating, dual-signal biosensor for carbamate pesticides, integrating all three amplification strategies.
Protocol 1: Synthesis of Fluorescent Gold Nanozymes (AuNEs)
Principle: Bovine Serum Albumin (BSA) serves as both a reducing and a stabilizing agent to form fluorescent gold nanoclusters with peroxidase-like activity [52].
Materials:
- Chloroauric acid (HAuCl₄)
- Bovine Serum Albumin (BSA)
- Sodium hydroxide (NaOH)
- Ultrapure water
Procedure:
- Prepare an aqueous solution of HAuCl₄ (10 mM) and BSA (50 mg/mL) in ultrapure water.
- Rapidly mix the HAuCl₄ and BSA solutions under vigorous stirring at a molar ratio optimized for maximum fluorescence (typically 1:100 - 1:200 HAuCl₄:BSA).
- Adjust the pH of the reaction mixture to 10-12 using 1M NaOH to facilitate the reduction of Au³⁺ to Au⁰ by BSA.
- Incubate the reaction mixture at 37°C for 12-24 hours. The color of the solution will change from light yellow to deep brown, indicating the formation of AuNEs.
- Purify the synthesized AuNEs by dialysis or centrifugation to remove unreacted precursors.
- Characterize the AuNEs using Transmission Electron Microscopy (TEM) for size and morphology, UV-Vis spectroscopy for absorption, and fluorescence spectroscopy for emission properties.
Protocol 2: Fabrication of the Biosensor and Pesticide Detection
Principle: This protocol integrates the AChE/CHO catalytic cascade with the dual-signal output (colorimetric and fluorometric) of the AuNEs. The presence of carbamate pesticides inhibits the cascade, quantitatively reducing both signals [52].
Materials:
- Biological Reagents: Acetylcholinesterase (AChE), Choline Oxidase (CHO), Acetylcholine (ACh)
- Nanozymes: Synthesized BSA-AuNEs (from Protocol 1)
- Chromogen/Substrates: 3,3',5,5'-Tetramethylbenzidine (TMB), Ferrous Chloride (FeCl₂)
- Buffer: Phosphate Buffered Saline (PBS, 0.1 M, pH 7.4)
- Samples: Carbamate pesticide standards (e.g., carbofuran, aldicarb) and prepared fruit extracts.
Procedure: Part A: Enzymatic Cascade and H₂O₂ Generation
- Prepare the cascade reaction mixture in a 1 mL cuvette or microcentrifuge tube:
- PBS Buffer (0.1 M, pH 7.4): 800 µL
- AChE (1 U/mL): 50 µL
- CHO (1 U/mL): 50 µL
- Varying concentrations of carbamate pesticide standard or fruit sample extract: 50 µL
- Incubate the mixture at 37°C for 10 minutes.
- Add 50 µL of Acetylcholine (ACh, 10 mM) to initiate the cascade reaction. Incubate for another 20 minutes at 37°C. The H₂O₂ produced in this step is proportional to the uninhibited enzyme activity.
Part B: Dual-Signal Detection
- Split the reaction mixture from Part A into two equal aliquots (~500 µL each) for parallel colorimetric and fluorometric analysis.
Detection Workflow:
Data Analysis and Calibration
- Calibration Curve: Plot the absorbance (colorimetric) and relative fluorescence intensity (fluorometric) against the logarithm of the carbamate pesticide concentration. The signals should decrease with increasing pesticide concentration due to enzyme inhibition.
- Quantification: Use the linear range of the calibration curve to interpolate the concentration of pesticides in unknown fruit samples.
- Self-Calibration: The mutual verification between the two orthogonal signals (colorimetric and fluorometric) provides an internal self-correction mechanism, enhancing the anti-interference capability against complex fruit matrices [52].
Table 2: Typical Performance Metrics for the Dual-Signal Carbamate Detection
| Pesticide (Example) | Detection Mode | Linear Range (ng/mL) | Limit of Detection (LOD, ng/mL) | Maximum Residue Limit (MRL) Reference [52] |
|---|---|---|---|---|
| Carbofuran | Colorimetric | 10 - 200 | 2.3 - 8.9 | 20 ppb (China) |
| Carbofuran | Fluorometric | 10 - 200 | 4.9 - 9.7 | 20 ppb (China) |
| Methomyl | Colorimetric | 10 - 200 | 2.3 - 8.9 | 100-200 ppb (China) |
| Methomyl | Fluorometric | 10 - 200 | 4.9 - 9.7 | 100-200 ppb (China) |
| Aldicarb | Colorimetric | 10 - 200 | 2.3 - 8.9 | 20 ppb (China) |
| Aldicarb | Fluorometric | 10 - 200 | 4.9 - 9.7 | 20 ppb (China) |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Sensor Development
| Reagent / Material | Function / Role in Assay | Example & Notes |
|---|---|---|
| Acetylcholinesterase (AChE) | Primary recognition element; inhibited by target pesticides. | Source from electric eel or recombinant; activity and purity are critical. |
| Choline Oxidase (CHO) | Second enzyme in cascade; generates detectable H₂O₂. | Used in conjunction with AChE to create the amplification cascade. |
| Nanozymes | Signal transducer and amplifier; replaces natural enzymes like HRP. | BSA-AuNEs [52], Fe₇S₈ NFs [53], Pd@Pt NPs [55]. Offer superior stability. |
| Chromogenic Substrate | Provides visual/absorbance signal upon oxidation. | TMB is most common, yielding a blue product (oxTMB) measurable at 652 nm [52] [53]. |
| Interdigitated Array (IDA) Electrode | Platform for redox cycling amplification. | Features closely spaced microelectrodes for generator-collector operation [56] [57]. |
| Buffer Systems (PBS) | Maintains optimal pH and ionic strength for enzymatic activity. | 0.1 M Phosphate Buffered Saline (PBS), pH 7.4, is typical for AChE/CHO [52]. |
The integration of nanozymes, redox cycling, and catalytic cascades presents a powerful toolkit for advancing electrochemical biosensors. The protocols outlined here for detecting fruit pesticide residues demonstrate how these strategies can be combined to create highly sensitive, robust, and self-validating analytical platforms. The move towards organic-dominated nanozymes and the incorporation of smartphone-based readout systems, as highlighted in the provided research, will further enhance the portability, sustainability, and applicability of these methods for real-world food safety monitoring [53] [54] [14]. These signal amplification strategies hold immense potential for transforming the detection of not only pesticides but a wide array of analytes in complex biological and environmental samples.
The detection of pesticide residues in fruits is a critical component of ensuring food safety and protecting public health. Within this field, electrochemical biosensors have emerged as a promising technology due to their potential for rapid, sensitive, and on-site analysis. The performance of these biosensors is profoundly influenced by the biochemical conditions under which they operate. This application note provides detailed protocols for the systematic optimization of three fundamental assay parameters—pH, ionic strength, and incubation time—specifically for electrochemical biosensors targeting pesticide residues in fruit matrices. Proper optimization is essential for maximizing the analytical sensitivity, specificity, and overall reliability of the biosensing platform, which integrates biological recognition elements with electrochemical transducers to convert a biological response into a quantifiable electrical signal [14] [58].
The inherent complexity of fruit samples, which contain various interfering compounds such as organic acids, sugars, and pigments, makes the optimization process particularly crucial. These matrix components can affect the activity of the biorecognition element (e.g., enzymes, aptamers, antibodies) and the efficiency of the electron transfer process at the electrode surface [14]. Furthermore, the binding affinity and reaction kinetics between the bioreceptor and the target pesticide analyte are highly dependent on the physicochemical environment. Therefore, a methodical approach to optimizing the assay milieu is not merely a procedural step but a foundational requirement for developing a robust and accurate analytical method fit for purpose in research and potential commercial application.
Theoretical Background
The Role of Key Assay Parameters
The analytical signal in an electrochemical biosensor is the culmination of a complex interplay between the biorecognition event and the subsequent electrochemical transduction. The following parameters are fundamental to this process:
- pH: The pH of the assay buffer directly impacts the ionization state of amino acid residues in the active site of enzyme-based biosensors, thereby governing catalytic activity and stability. For immunosensors and aptasensors, pH influences the three-dimensional structure and binding affinity of antibodies and aptamers. Deviation from the optimal pH range can lead to irreversible denaturation and a significant loss of signal. Moreover, pH can affect the charge on the electrode surface and the electrochemical behavior of reporter molecules [14] [58].
- Ionic Strength: The ionic concentration of the solution influences the electrochemical double layer at the electrode-solution interface, which can either facilitate or hinder electron transfer kinetics. From a biological perspective, ionic strength is critical for stabilizing the structure of biomolecules. For instance, it shields the negative charges on the phosphate backbone of DNA aptamers, affecting their folding and binding conformation towards target pesticides. However, excessively high ionic strength can also mask electrostatic interactions that are essential for specific binding events [14].
- Incubation Time: This parameter dictates the duration allowed for the specific binding reaction between the biorecognition element and the target pesticide to reach equilibrium. An insufficient incubation time results in an incomplete reaction and a sub-optimal signal, whereas an excessively long incubation time reduces analysis throughput and may increase non-specific binding, leading to a higher background signal [58].
Biosensor Signaling Mechanisms
A foundational signaling mechanism for pesticide detection, particularly for organophosphates and carbamates, is enzyme inhibition. The following diagram illustrates the operational principle of a common acetylcholinesterase (AChE)-based electrochemical biosensor.
Diagram 1: Biosensor signaling via enzyme inhibition. Acetylcholinesterase (AChE) is immobilized on the electrode. Its substrate, acetylthiocholine, is converted to thiocholine, which generates an electrochemical signal (e.g., via oxidation). Pesticide binding inhibits AChE, reducing the product formation and causing a measurable signal drop proportional to pesticide concentration [14] [58].
Experimental Protocols
Reagent Preparation
Note: Use high-purity deionized water (≥18 MΩ·cm) and analytical grade reagents for all preparations.
- Phosphate Buffered Saline (PBS), 10x Stock (1 L):
- Weigh 80.0 g of NaCl, 2.0 g of KCl, 14.4 g of Na₂HPO₄, and 2.4 g of KH₂PO₄.
- Dissolve in 800 mL of deionized water.
- Adjust the pH to 7.4 using either 1 M HCl or 1 M NaOH.
- Make up the final volume to 1 L with deionized water. Sterilize by autoclaving or filtration (0.22 µm). This stock can be stored at 4°C for up to 6 months.
- Working Buffer (from 10x PBS):
- Prepare 1x PBS by diluting the 10x stock ten-fold with deionized water.
- This 1x PBS (pH 7.4) serves as the baseline buffer for all optimization experiments.
Sample Preparation (Fruit Matrices)
The sample preparation protocol is critical for minimizing matrix effects. The following workflow outlines a streamlined procedure based on the QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) approach, which is widely adopted for complex matrices like fruits [5] [59].
Diagram 2: Workflow for fruit sample preparation. Homogenizing the fruit in a solvent like acetonitrile extracts pesticides and other components. Salt-induced partitioning helps separate the organic phase from water and solids. The final extract must be diluted into a compatible aqueous buffer (e.g., PBS) for biosensor analysis [5] [59].
Optimization Procedure: A Systematic Approach
A univariate or multivariate (e.g., Design of Experiments) approach can be employed. The protocol below describes a comprehensive univariate method.
- Apparatus: Electrochemical workstation (e.g., potentiostat), three-electrode system (working, reference, counter), magnetic stirrer, pH meter, analytical balance.
- Biosensor Platform: The protocol assumes a ready-to-use biorecognition layer (e.g., AChE, antibody, or aptamer) immobilized on the working electrode.
Protocol Steps:
pH Optimization:
- Prepare a series of 1x PBS buffers with identical ionic strength but varying pH levels (e.g., 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5) using 1 M HCl or NaOH.
- Reconstitute the pesticide standard and the biosensor in each pH buffer.
- Perform the assay under otherwise identical conditions (e.g., 25°C, 15 min incubation).
- Measure the electrochemical response (e.g., current for enzyme-based sensors, impedance change for label-free sensors).
- Plot the signal-to-noise ratio (SNR) against pH to identify the optimum.
Ionic Strength Optimization:
- Prepare a series of buffers at the optimal pH (determined in Step 1) with varying ionic strengths. This can be achieved by diluting the 10x PBS stock to different concentrations (e.g., 0.1x, 0.5x, 1x, 2x) or by adding precise amounts of NaCl (e.g., 0 mM, 50 mM, 100 mM, 150 mM, 200 mM) to a low-salt buffer.
- Perform the assay with the biosensor in each buffer.
- Plot the SNR against ionic strength (or NaCl concentration) to identify the optimum.
Incubation Time Optimization:
- Using the optimal pH and ionic strength buffer, perform the assay with varying incubation times (e.g., 1, 5, 10, 15, 20, 30 minutes).
- Ensure all other conditions (temperature, concentration) remain constant.
- Measure the electrochemical response at each time point.
- Plot the signal intensity against time. The optimal time is typically at or near the signal plateau, indicating reaction equilibrium.
Data Presentation and Analysis
The following table consolidates typical optimal ranges for key assay parameters based on current literature for various biosensor types. These ranges should be used as a starting point for experimental design.
Table 1: Typical Optimal Ranges for Key Assay Parameters in Pesticide Biosensors
| Parameter | Typical Optimal Range | Key Considerations & Impact of Deviation |
|---|---|---|
| pH | 7.0 - 7.8 | - Enzyme-based sensors (AChE): Maximal activity in neutral pH. Lower pH causes protonation, higher pH leads to denaturation [14].- Immunosensors/Aptasensors: Dependent on bioreceptor's isolectric point. Affects binding affinity and complex stability. |
| Ionic Strength | 100 - 150 mM NaCl | - Stabilizes biomolecule structure and folding (critical for aptamers) [14].- Low strength: Can cause non-specific adsorption and unstable baseline.- High strength: Can mask electrostatic interactions, reducing binding affinity and signal. |
| Incubation Time | 10 - 20 minutes | - Time for binding reaction to reach ~90% of equilibrium [58].- Too short: Low signal, poor sensitivity.- Too long: Increased risk of non-specific binding, reduced throughput. |
Exemplary Optimization Dataset
The table below presents a hypothetical dataset from a systematic optimization experiment for an aptamer-based electrochemical biosensor detecting imidacloprid in apple extracts.
Table 2: Exemplary Dataset from Optimization of an Aptamer-Based Biosensor
| Condition Tested | Parameter Value | Normalized Signal (%) | Signal-to-Noise Ratio | Recommended Optimal Value |
|---|---|---|---|---|
| pH | 6.5 | 65 | 8.5 | |
| 7.0 | 88 | 12.1 | ||
| 7.5 | 100 | 15.2 | pH 7.5 | |
| 8.0 | 92 | 13.0 | ||
| 8.5 | 75 | 9.1 | ||
| Ionic Strength (NaCl, mM) | 50 | 70 | 7.8 | |
| 100 | 100 | 15.2 | 100 mM | |
| 150 | 95 | 13.5 | ||
| 200 | 80 | 10.1 | ||
| Incubation Time (min) | 5 | 55 | 6.5 | |
| 10 | 85 | 11.8 | ||
| 15 | 100 | 15.2 | 15 min | |
| 20 | 98 | 14.9 | ||
| 30 | 99 | 14.8 |
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Biosensor Development
| Item | Function/Application in Biosensor Development |
|---|---|
| Phosphate Buffered Saline (PBS) | Universal buffer for maintaining physiological pH and osmolarity during bioreceptor immobilization and assay steps [14]. |
| Acetylcholinesterase (AChE) | Key enzyme for organophosphate and carbamate pesticide detection; inhibition by these pesticides provides the basis for the analytical signal [14] [58]. |
| Nucleic Acid Aptamers | Synthetic single-stranded DNA/RNA molecules acting as bioreceptors; selected for high affinity to specific pesticides (e.g., acetamiprid, imidacloprid) [14] [58]. |
| Monoclonal Antibodies | Biological recognition elements providing high specificity for target pesticides in immunosensor configurations [14] [58]. |
| Electrochemical Redox Probes | Molecules such as ([Fe(CN)_6]^{3-/4-}) used to probe electron transfer efficiency at the electrode surface, often measured via Electrochemical Impedance Spectroscopy (EIS) [58]. |
| QuEChERS Extraction Kits | Standardized kits for efficient extraction and clean-up of pesticide residues from complex fruit matrices, minimizing interferents in the final analysis [5] [59]. |
Troubleshooting and Technical Notes
- Problem: High Background Signal.
- Potential Cause: Non-specific adsorption of matrix components to the electrode or bioreceptor surface.
- Solution: Include a blocking agent (e.g., 1% BSA, casein) in the buffer after bioreceptor immobilization. Optimize wash steps post-incubation. Further dilution of the sample extract may be necessary.
- Problem: Low Signal Intensity.
- Potential Cause 1: Sub-optimal pH or ionic strength inactivates the bioreceptor.
- Solution: Re-verify the optimal pH and ionic strength using a standard solution of the target pesticide.
- Potential Cause 2: Insufficient incubation time.
- Solution: Perform a full time-course experiment to confirm the signal has reached a plateau.
- Problem: Poor Reproducibility.
- Potential Cause: Inconsistent buffer preparation or temperature fluctuations during incubation.
- Solution: Precisely prepare all buffers from a single, concentrated stock. Use a temperature-controlled environment (e.g., water bath) for all incubation steps.
- Note on Matrix Effects: Always validate the optimized assay conditions using the target fruit matrix (e.g., apple, grape) spiked with known concentrations of the pesticide. The complex composition of fruits can significantly alter the optimal parameters established in pure buffer systems [14] [5].
Ensuring Sensor Stability, Regeneration, and Shelf-Life
Electrochemical biosensors represent a powerful tool for the rapid detection of pesticide residues on fruits, aligning with the growing demands of precision agriculture and food safety monitoring [10] [60]. A critical determinant for the transition of these biosensors from research laboratories to widespread field deployment is their robustness, characterized by long-term stability, reliable regeneration capabilities, and a predictable shelf-life. The biological and chemical components of biosensors are prone to ageing, defined as a decrease in signal response over time, which can undermine analytical accuracy and user confidence [61]. This application note provides detailed protocols and data-driven insights to help researchers systematically evaluate and enhance these critical performance parameters, ensuring the development of commercially viable sensing platforms.
Quantitative Data on Biosensor Stability
The following tables consolidate key stability characteristics and parameters essential for planning and interpreting biosensor ageing studies.
Table 1: Biosensor Stability Characteristics and Testing Methods
| Stability Characteristic | Description | Common Testing Method | Key Influencing Factors |
|---|---|---|---|
| Shelf-Life | Long-term stability during storage before use. | Thermally accelerated ageing at elevated temperatures [61]. | Storage temperature, immobilization matrix, humidity, biological element stability [61]. |
| Operational Stability | Stability during continuous use in analysis. | Continuous electrochemical interrogation in buffer or sample matrix [61]. | Temperature, applied potential, matrix effects (e.g., fouling), analyte concentration. |
| Reusability | Ability to be used multiple times after regeneration. | Repeated measurement-regeneration cycles [61] [62]. | Regeneration protocol efficiency, sensor surface fouling, handling physical damage [61]. |
Table 2: Key Parameters from Stability and Regeneration Studies
| Parameter | Value / Range | Experimental Context | Reference |
|---|---|---|---|
| Shelf-Life Prediction | Linear ageing model more suitable than exponential Arrhenius model | Model comparison for glucose oxidase biosensors [61]. | [61] |
| Accelerated Ageing Duration | ~4 days for shelf-life determination; <24 hours for continuous use stability | Thermally accelerated ageing protocol [61]. | [61] |
| Signal Retention | >75% original signal after 50 days aqueous storage | E-DNA sensor with flexible trihexylthiol anchor [62]. | [62] |
| Regeneration Efficiency | >91% signal recovery after 30s wash in deionized water | E-DNA sensor platform [62]. | [62] |
| Detection Limit | Nanomolar range (high ppt) for dichlorvos | On-glove inhibition biosensor for fruit peels [10]. | [10] |
Experimental Protocols
Protocol for Thermally Accelerated Ageing to Predict Shelf-Life
This protocol provides a rapid method to estimate the long-term shelf-life of electrochemical biosensors, based on established models [61].
- Principle: Ageing is accelerated at elevated temperatures, and a linear model is used to extrapolate stability at standard storage temperatures.
- Materials:
- Fabricated biosensors (e.g., screen-printed electrodes).
- Temperature-controlled incubators (e.g., set at 4°C, 25°C, 37°C, 50°C).
- Standard analyte solutions at known concentrations.
- Electrochemical workstation (e.g., potentiostat).
- Appropriate storage buffer (e.g., phosphate-buffered saline).
- Procedure:
- Baseline Measurement: For each sensor batch, perform electrochemical measurements (e.g., chronoamperometry, cyclic voltammetry) using a standard analyte solution to establish the initial signal (S₀).
- Accelerated Ageing: Divide the sensors into groups and store each group in sealed containers at different elevated temperatures (e.g., 4°C, 25°C, 37°C, 50°C) for a defined period (e.g., 4 days) [61].
- Periodic Sampling: At predetermined time intervals, remove a subset of sensors from each temperature condition and measure the signal (Sₜ) using the same standard method from step 1.
- Data Analysis:
- For each temperature, plot the normalized signal (Sₜ/S₀) against time.
- Fit a linear regression to the data for each temperature to determine the degradation rate.
- Use the linear model to extrapolate the time required for the signal to drop to a predefined threshold (e.g., 90% of S₀) at the desired storage temperature (e.g., 4°C).
Protocol for Evaluating Continuous Use Stability and Reusability
This procedure assesses sensor performance under repeated operational and regeneration cycles, which is critical for applications requiring multiple measurements.
- Principle: The sensor is subjected to consecutive cycles of analyte measurement followed by a regeneration step, monitoring the signal fidelity over time.
- Materials:
- Functionalized biosensor.
- Electrochemical workstation.
- Analyte solutions at relevant concentrations (e.g., pesticide standards).
- Regeneration solution (e.g., deionized water, mild acid/base, or specific chelating agents) [62].
- Test matrix (e.g., buffer, simulated fruit peel extract).
- Procedure:
- Initial Characterization: Measure the sensor's response in the test matrix without analyte (background signal) and with a known concentration of target analyte (S_initial).
- Stability & Reusability Cycle:
- Step A (Measurement): Incubate the sensor with the analyte solution for a fixed time (e.g., ~5 minutes [62]) and record the electrochemical signal.
- Step B (Regeneration): Apply the regeneration protocol. For example, rinse the sensor with deionized water for 30 seconds [62] or incubate in a regeneration solution.
- Step C (Signal Recovery): Measure the sensor's signal in the test matrix without analyte to confirm signal recovery.
- Repeat the cycle (Steps A-C) for the desired number of times (n cycles).
- Data Analysis:
- Plot the measured signal from Step A and the background signal from Step C against the cycle number.
- Calculate the signal retention after n cycles as (Scyclen / Sinitial) × 100%.
- The reusability is determined by the number of cycles before the signal degrades below an acceptable threshold (e.g., <80% of Sinitial).
Visualization of Workflows and Architectures
Biosensor Ageing Study Workflow
The following diagram outlines the logical workflow for conducting a comprehensive biosensor stability assessment.
Stable SAM-Based Biosensor Architecture
This diagram illustrates the design of an electrochemical biosensor utilizing a stable self-assembled monolayer (SAM) anchor, a key strategy for improving shelf-life.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagents for Biosensor Fabrication and Stability Testing
| Item | Function / Application | Key Considerations |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, miniaturized working electrode platform. Ideal for decentralized analysis [10] [61]. | Carbon, gold, or platinum working electrodes. Surface pre-treatment may be required. |
| Thiol-Based Anchors (e.g., Trihexylthiol) | Form self-assembled monolayers (SAMs) on gold to immobilize bioreceptors. Enhance stability versus monothiols [62]. | Flexibility of anchor impacts packing and stability. Rigid anchors may offer less improvement. |
| Prussian Blue | High-efficiency electrocatalyst; used as a redox mediator in biosensors [10] [61]. | Often called "artificial peroxidase." Stability is crucial for overall sensor performance [61]. |
| Butyrylcholinesterase (BChE) Enzyme | Biorecognition element in organophosphorus pesticide biosensors [10]. | Enzyme inhibition by pesticides is the detection mechanism. Enzyme stability dictates sensor lifetime. |
| 6-Mercapto-1-hexanol (MCH) | Backfilling agent in SAMs to displace non-specific adsorption and improve probe orientation [62]. | Reduces non-specific binding and enhances electron transfer efficiency. |
| Phosphate Buffered Saline (PBS) | Standard storage and measurement buffer. Maintains pH and ionic strength. | pH and ionic strength can significantly affect biosensor response, especially immunosensors [20]. |
Addressing Common Interferents and False-Positive Signals
Electrochemical biosensors are powerful tools for detecting pesticide residues in fruit, but their accuracy can be compromised by various interferents present in complex fruit matrices. These interferents can cause false-positive signals, potentially leading to incorrect conclusions about pesticide contamination levels. Understanding and mitigating these interferents is therefore crucial for developing reliable analytical protocols for food safety monitoring [63] [10].
The fundamental operation of an electrochemical biosensor involves a biological recognition element (such as an enzyme) interacting with the target analyte, generating an electrochemical signal that is transduced and measured. Common interferents in fruit samples include naturally occurring compounds such as ascorbic acid, phenolic compounds, flavonoids, and sugars, which can either undergo direct redox reactions at the electrode surface or inhibit the biological recognition element [14]. This application note provides a structured framework for identifying, characterizing, and mitigating these interferents to enhance the reliability of pesticide detection in fruit samples.
Common Interferents and Mitigation Strategies
Table 1: Common Interferents in Electrochemical Biosensors for Fruit Pesticide Detection
| Interferent Category | Specific Examples | Source in Fruit Matrices | Interference Mechanism | Mitigation Strategies |
|---|---|---|---|---|
| Electroactive Compounds | Ascorbic acid, catechols, uric acid | Naturally occurring antioxidants in fruits (e.g., citrus, apples) | Direct oxidation at electrode potential, generating non-specific current | Use permselective membranes (Nafion), electrode surface passivation, potential cycling cleaning |
| Enzyme Inhibitors | Heavy metals (Pb, Cd), fluoride | Environmental contamination, some tea varieties [14] | Non-competitive inhibition of enzyme activity (e.g., acetylcholinesterase) | Sample dilution, chelating agents, use of enzyme inhibitors in control experiments |
| Protein-Binding Compounds | Polyphenols, tannins | Grapes, berries, pomegranates | Non-specific binding to bioreceptor, fouling electrode surface | Surface blocking agents (BSA, casein), filtration, solid-phase extraction |
| Surface-Active Compounds | Lipids, surfactants | Fruit waxes, post-harvest treatments | Adsorption on electrode surface, modifying electron transfer kinetics | Electrode polishing, surfactant additives, pulsed electrochemical techniques |
| Structural Analogues | Other organophosphorus compounds | Multiple pesticide applications | Cross-reactivity with biorecognition element | Use of more specific bioreceptors (aptamers, MIPs), multidimensional sensing approaches |
Experimental Protocol for Interferent Evaluation
Materials and Reagents
Table 2: Essential Research Reagent Solutions
| Reagent/Material | Function/Application | Preparation/Specification |
|---|---|---|
| Acetylcholinesterase (AChE) | Biological recognition element for organophosphorus pesticides | 0.5 U/mL in phosphate buffer (pH 7.4), aliquot and store at -20°C |
| Prussian Blue/Carbon Black nanocomposite | Electron-transfer mediator for signal amplification | Synthesize as in [10], suspend in deionized water (1 mg/mL) |
| Screen-printed carbon electrodes (SPCEs) | Transducer platform | Commercially sourced or fabricated in-house with Ag/AgCl reference |
| Phosphate Buffered Saline (PBS) | Electrochemical baseline medium | 0.1 M, pH 7.4, containing 0.1 M KCl as supporting electrolyte |
| Dichlorvos standard | Target analyte (organophosphorus pesticide) | Prepare stock solution (1000 ppm in methanol), store at 4°C |
| Ascorbic acid solution | Model interferent for validation | Prepare daily in PBS (0.1 M) from solid form |
| Nafion permeslective membrane | Interferent exclusion layer | 0.5% solution in lower aliphatic alcohols |
Step-by-Step Methodology
Protocol: Assessment of Ascorbic Acid Interference in Organophosphorus Pesticide Detection
Principle: This protocol evaluates the extent of interference from ascorbic acid, a common electroactive compound in fruits, during the detection of organophosphorus pesticides using an acetylcholinesterase-based biosensor. The approach follows the biosensor design principles outlined in [10] with specific modifications for interferent analysis.
Procedure:
- Biosensor Preparation:
- Modify screen-printed carbon electrodes with 5 μL of Prussian Blue/Carbon Black nanocomposite suspension.
- Allow to dry at room temperature for 30 minutes.
- Immobilize acetylcholinesterase by depositing 3 μL of enzyme solution (0.5 U/mL) and allowing cross-linking with 1% glutaraldehyde vapor for 15 minutes.
- For interferent exclusion, apply 2 μL of 0.5% Nafion solution and spin-coat at 2000 rpm for 30 seconds.
Control Measurement (Without Interferent):
- Immerse the prepared biosensor in 10 mL PBS containing 50 nM dichlorvos (target pesticide).
- Record differential pulse voltammograms from -0.2V to +0.6V vs. Ag/AgCl reference.
- Measure peak current at +0.35V as reference signal.
Interferent Challenge:
- Transfer the same biosensor to 10 mL PBS containing 50 nM dichlorvos and 100 μM ascorbic acid.
- Record voltammograms using identical parameters.
- Measure peak current at +0.35V.
Data Analysis:
- Calculate signal enhancement (%) = [(Iinterferent - Icontrol)/I_control] × 100
- A signal enhancement >5% indicates significant interference requiring mitigation.
Validation with Real Sample:
- Extract pesticide from fruit peel (apple, orange) using methanol:water (80:20) solution.
- Analyze with and without standard addition of known pesticide concentration.
- Compare results with and without Nafion modification to confirm interferent suppression.
Diagram 1: Experimental protocol for interferent evaluation (Title: Interferent Test Workflow)
Advanced Interference Mitigation Approaches
Multi-phase Electron Transfer Engineering
Recent advances in electron transfer control offer sophisticated approaches to interference mitigation. Cascade-responsive microsystems with multi-phase electron transfer reactions can significantly enhance sensing reliability by minimizing background interference through several mechanisms: avoiding target information loss, implementing signal amplification strategies, and actively removing background interference [64].
Table 3: Advanced Signal Enhancement and Interference Suppression Techniques
| Technique | Principle | Implementation in Pesticide Detection |
|---|---|---|
| Constrained Electron Transfer Cascades | Spatial confinement of redox reactions | Homogeneous electrochemical sensors with localized signal generation away from interfering species |
| Interfacial Collision Electrochemistry | Transient signals from discrete binding events | Discrimination based on event frequency and amplitude rather than steady-state current |
| Multi-phase Reaction Control | Compartmentalization of reaction steps | Microfluidic separation of sample matrix from detection zone |
| Dynamic Feature Extraction | Analysis of kinetic parameters rather than endpoint measurements | Time-dependent signal processing to distinguish specific binding from non-specific interactions |
Diagram 2: Advanced interference mitigation strategy (Title: Signal Cleaning Process)
Validation Framework for Fruit Pesticide Detection
Establishing a robust validation framework is essential for confirming that interference mitigation strategies are effective. The following protocol outlines a comprehensive approach:
Protocol: Comprehensive Biosensor Validation Against Matrix Effects
Selectivity Profile:
- Challenge the biosensor with individual fruit matrix components (ascorbic acid, fructose, malic acid, common flavonoids)
- Establish maximum tolerable concentration for each potential interferent
Standard Addition Method:
- Spike known pesticide concentrations into different fruit extracts (apple, orange, grape)
- Calculate recovery percentages (acceptable range: 85-115%)
- Compare results with LC-MS/MS reference method
Cross-reactivity Assessment:
- Test biosensor against structurally related pesticides
- Quantify cross-reactivity as percentage relative to target pesticide
Long-term Stability:
- Monitor biosensor response over 4-week period with periodic testing
- Store under recommended conditions between measurements
Implementing these protocols will significantly enhance the reliability of electrochemical biosensors for detecting pesticide residues in fruit, minimizing false-positive signals and ensuring accurate results for food safety monitoring.
Benchmarking Biosensor Efficacy: Validation Against Gold Standards and Regulatory Alignment
The increasing use of pesticides in agricultural production has introduced significant health concerns due to pesticide residue accumulation in fruits and vegetables [5] [18]. Electrochemical biosensors present attractive alternatives to conventional analytical techniques like LC-MS/MS and GC-MS/MS, offering specificity, sensitivity, speed, and potential for on-site analysis [65]. However, to ensure reliable field deployment for fruit pesticide residue detection, rigorous validation of key analytical figures of merit is essential. This protocol details the establishment of Limit of Detection (LOD), Limit of Quantification (LOQ), linearity, and reproducibility for electrochemical biosensors within the context of fruit pesticide residue analysis, providing a standardized framework for researchers and scientists.
Defining the Core Analytical Figures of Merit
Limit of Detection (LOD) and Limit of Quantification (LOQ)
The Limit of Detection (LOD) is the lowest concentration of an analyte that can be reliably distinguished from the absence of that analyte. For biosensors, the LOD is defined as the concentration where the signal (S) is three times greater than the noise (N), or equivalently, when the signal is greater than three standard deviations of the blank measurement (S > 3σ) [66].
The Limit of Quantification (LOQ) is the lowest concentration of an analyte that can be quantitatively determined with suitable precision and accuracy. The LOQ is defined as the concentration where the signal is ten times greater than the noise (S/N > 10), or when the signal is greater than ten times the standard deviation (S > 10σ) [66]. In practical pesticide residue analysis, the LOQ values must be much smaller than the Maximum Residue Levels (MRLs) established by regulatory bodies such as the European Union [67] [68].
Linearity
Linearity refers to the ability of a biosensor to produce a signal that is directly proportional to the concentration of the analyte within a specified range [65]. This range is known as the Analytical Range or the linear dynamic range. It is the interval between the upper and lower concentrations where the sensor has been demonstrated to be precise and accurate [66]. The linearity is typically evaluated using the correlation coefficient (R²) of the calibration curve, with values higher than 0.99 considered indicative of good linearity [67] [68].
Reproducibility
Reproducibility describes the precision of the biosensor, indicating the closeness of agreement between independent results obtained under stipulated conditions. It is often expressed as the Relative Standard Deviation (RSD) of repeated measurements [65]. For a method to be considered acceptably precise, the RSD values should generally be less than 20% [67] [68]. Related to reproducibility is Signal Drift, which describes the stability of a sensor's output signal when all conditions are fixed. Minimizing drift is crucial for maintaining reproducibility over time [66].
Table 1: Definitions and Evaluation Criteria for Key Analytical Figures of Merit
| Figure of Merit | Definition | Common Evaluation Criterion | Importance in Pesticide Detection |
|---|---|---|---|
| Limit of Detection (LOD) | Lowest concentration distinguishable from blank | S/N > 3 or S > 3σ [66] | Determines ability to detect trace residues |
| Limit of Quantification (LOQ) | Lowest concentration quantifiable with precision | S/N > 10 or S > 10σ [66] | Must be below MRLs for regulatory compliance [67] |
| Linearity | Proportionality of signal to analyte concentration | R² > 0.99 [67] [68] | Ensures accurate quantification across working range |
| Reproducibility | Precision of repeated measurements | RSD < 20% [67] [68] | Guarantees reliability of results across different operators and instruments |
Experimental Protocols for Determination
This section provides detailed methodologies for establishing the core figures of merit, using an example of an inhibition-based electrochemical biosensor for organophosphorus pesticides [10].
Reagent Preparation and Biosensor Fabrication
Materials:
- Transducer: Screen-printed electrodes (SPEs) [10]
- Biorecognition Element: Butyrylcholinesterase (BChE) enzyme [10]
- Electrochemical Probe: Prussian blue and Carbon black nanocomposite [10]
- Substrate: Butyrylthiocholine
- Analyte: Pesticide standards (e.g., Dichlorvos)
- Fruit Matrices: Apple and orange peels [10]
Biosensor Fabrication Protocol:
- Electrode Modification: Drop-cast a precise volume (e.g., 5-10 µL) of the bio-hybrid probe (Prussian blue, Carbon black, and BChE) onto the working electrode of the SPE.
- Immobilization: Allow the probe to dry and immobilize at room temperature for a set period (e.g., 1 hour).
- Curing: Place the modified electrodes in a desiccator at 4°C overnight to stabilize the biocomposite layer.
- Quality Control: Perform an initial electrochemical characterization (e.g., Cyclic Voltammetry) in a buffer solution to verify successful modification and consistent baseline performance across all sensors in a batch.
Protocol for Determining LOD and LOQ
Calibration Curve Generation:
- Prepare a series of standard solutions of the target pesticide (e.g., Dichlorvos) in a suitable buffer across a concentration range expected in real samples (e.g., 0.1 nM to 100 µM).
- For each concentration, incubate the biosensor for a fixed time (e.g., 10 minutes).
- Measure the amperometric response (e.g., current decrease due to enzyme inhibition) under optimized conditions (fixed voltage, pH, temperature).
- Perform each measurement in triplicate (n=3) to assess variability.
Signal and Noise Measurement:
- Record the signals for each standard concentration.
- Measure the noise (standard deviation, σ) from the response of multiple blank (pesticide-free) samples (n ≥ 10).
Calculation:
- LOD = 3.3 × (σ / S), where σ is the standard deviation of the blank response and S is the slope of the calibration curve in the low concentration region.
- LOQ = 10 × (σ / S).
- Confirm that the calculated LOD is in the nanomolar range (high ppt) for relevance to regulatory limits [10].
Protocol for Establishing Linearity and Analytical Range
- Data Collection: Use the data from the calibration curve generated in Section 3.2.
- Linear Regression Analysis:
- Plot the measured signal (y-axis) against the pesticide concentration (x-axis).
- Perform a least-squares linear regression analysis on the data points within the suspected linear range.
- Calculate the correlation coefficient (R²). The range is considered linear if R² ≥ 0.99 [68].
- Analytical Range Definition: The lower end of the analytical range is defined by the LOQ. The upper end is the highest concentration at which the sensor response remains linear and does not saturate.
Protocol for Assessing Reproducibility
Intra-assay Precision (Repeatability):
- Using the same biosensor and operator, analyze a single medium-level pesticide concentration (e.g., near the middle of the linear range) at least five times consecutively.
- Calculate the mean signal and the RSD for these measurements.
Inter-assay Precision (Reproducibility):
- Using multiple biosensors from the same fabrication batch (n ≥ 3), analyze the same medium-level pesticide concentration.
- Each measurement should be performed by a different operator if possible, using different reagent aliquots and on different days.
- Calculate the mean signal and the RSD for these measurements. An RSD of less than 10% is considered satisfactory for a portable biosensor [10].
Table 2: Example Experimental Data for an On-Glove Biosensor Detecting Dichlorvos [10]
| Figure of Merit | Experimental Result | Method / Notes |
|---|---|---|
| LOD | Nanomolar range (high ppt) | Lower than EU MRL for dichlorvos |
| LOQ | Nanomolar range | Suitable for regulatory compliance |
| Linearity | Demonstrated over relevant concentration range | R² value not explicitly stated, but implied by validation |
| Reproducibility (RSD) | < 10% | Satisfactory for a portable, on-glove system |
| Matrix Tested | Apple and orange peels | Direct analysis on fruit surfaces with minimal sample preparation |
Workflow and Signaling Pathway
The following diagram illustrates the complete experimental workflow for establishing the analytical figures of merit for an electrochemical biosensor, from preparation to data analysis.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents and Materials for Electrochemical Biosensor Development
| Reagent/Material | Function / Role | Example in Protocol |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, miniaturized transducer platform; enables mass fabrication and portability [10]. | On-glove biosensor for direct fruit peel analysis [10]. |
| Butyrylcholinesterase (BChE) Enzyme | Biorecognition element in inhibition-based biosensors; activity is inhibited by organophosphorus pesticides [10]. | Bio-hybrid probe for detecting Dichlorvos [10]. |
| Prussian Blue (PB) | High-efficiency electrocatalyst; mediates electron transfer, often used for low-potential detection of H₂O₂ in oxidase-based systems [10]. | Component of the nanocomposite to enhance sensitivity [10]. |
| Carbon Black (CB) | Nanomaterial that increases electrode surface area and enhances electron transfer kinetics, improving signal strength [10]. | Component of the nanocomposite to enhance signal [10]. |
| Butyrylthiocholine | Enzyme substrate; its hydrolysis product (thiocholine) is electrochemically detected, providing the measurable signal that decreases with pesticide inhibition [65]. | Substrate for BChE enzyme. |
| QuEChERS Kits | Sample preparation for complex matrices; not always needed for direct biosensor use but crucial for method validation against chromatographic standards [69] [67]. | Validating biosensor performance against LC-MS/MS or GC-MS/MS. |
Within analytical chemistry, liquid chromatography-tandem mass spectrometry (LC-MS/MS) and gas chromatography-mass spectrometry (GC-MS) represent two cornerstone techniques for the precise identification and quantification of chemical compounds. This application note provides a systematic comparison of their performance characteristics and practical trade-offs. The context for this discussion is a research project developing an electrochemical biosensor for detecting organophosphorus pesticides on fruit peels, where LC-MS/MS and GC-MS serve as the definitive reference methods for validating biosensor performance [10]. The selection between these techniques is pivotal and is governed by the chemical properties of the analytes, the complexity of the sample matrix, and the specific analytical requirements of the application.
Key Differences and Comparative Performance
The fundamental distinction between these techniques lies at the chromatographic stage. LC-MS/MS employs a liquid mobile phase to separate compounds, making it ideal for non-volatile, thermally labile, or high-molecular-weight compounds such as proteins, peptides, and many modern pesticides [70] [71]. In contrast, GC-MS utilizes a gaseous mobile phase and requires sample vaporization, making it exceptionally suited for volatile and semi-volatile compounds that can withstand the high temperatures of the analysis [70].
The following table summarizes the core performance metrics and trade-offs to guide method selection.
Table 1: Comparative Performance of LC-MS/MS and GC-MS for Pesticide Residue Analysis
| Characteristic | LC-MS/MS | GC-MS |
|---|---|---|
| Ideal Analyte Properties | Non-volatile, thermally labile, polar, high molecular weight [70] | Volatile, semi-volatile, thermally stable [70] |
| Common Ionization Techniques | Electrospray Ionization (ESI), Atmospheric Pressure Chemical Ionization (APCI) [70] [72] | Electron Ionization (EI), Chemical Ionization (CI) [70] [72] |
| Sample Preparation (for Pesticides) | Often requires extraction and cleanup; may need pH adjustment or buffer exchange [18] | Often requires extraction and cleanup; frequently necessitates chemical derivatization for non-volatile pesticides [70] [18] |
| Throughput | Moderate to High | Faster (with ultrafast GC techniques) [72] |
| Scope of Analytes | Broader range, including large and polar molecules [71] | Narrower range, limited to volatile/derivatized compounds [70] |
| Sensitivity | High (ppt/ppq levels with modern triple quads) [70] [72] | High (ppt levels possible) [70] [10] |
| Qualitative Libraries | Limited; spectra are instrument-dependent | Extensive; standardized EI spectral libraries available [70] |
| Instrument & Operational Costs | Generally higher initial investment and operational costs [70] | Generally lower initial investment and operational costs [70] |
Application in Pesticide Residue Analysis and Biosensor Validation
In the specific context of validating an electrochemical biosensor for fruit pesticides, both techniques are indispensable yet serve complementary roles [10] [18]. LC-MS/MS is exceptionally powerful for analyzing a wide range of pesticide classes, including carbamates, neonicotinoids, and many organophosphates, which are polar or have low thermal stability [18]. Its tandem mass spectrometry capability provides high specificity and confirmation power in complex food matrices like fruit extracts.
GC-MS, particularly GC-MS/MS, remains the gold standard for analyzing volatile pesticide classes, such as organochlorines (OCPs), pyrethroids, and some organophosphates [18]. Its high-resolution separation and robust spectral library matching make it a powerful confirmatory technique. A key practical consideration for GC-MS is that many non-volatile pesticides require a derivatization step to become volatile and thermally stable enough for analysis, which adds complexity and time to sample preparation [70].
The relationship between the novel biosensor method and these confirmatory techniques is foundational to method validation, as illustrated below.
Detailed Experimental Protocols
The following protocols are generalized for the analysis of pesticide residues in fruit peel extracts, designed to be adapted for specific instrument models and pesticide panels.
Protocol for LC-MS/MS Analysis of Pesticide Residues
Principle: Pesticides are extracted from the fruit matrix, separated via liquid chromatography based on polarity, ionized, and detected by tandem mass spectrometry using Multiple Reaction Monitoring (MRM) for high specificity and sensitivity [18].
Materials & Reagents:
- Fruit peel samples (e.g., apple, orange)
- LC-MS grade solvents: Acetonitrile, Methanol, Water
- Additives: Formic acid, Ammonium formate
- QuEChERS extraction kits or equivalent materials
- Internal Standards: Stable isotope-labeled pesticide standards
- Equipment: LC-MS/MS system (triple quadrupole recommended), analytical balance, centrifuge, vortex mixer
Procedure:
- Sample Preparation: Homogenize fruit peels. Weigh 10.0 ± 0.1 g of homogenate into a 50 mL centrifuge tube.
- Extraction: Add 10 mL of acetonitrile (1% formic acid) and appropriate internal standards. Shake vigorously for 1 minute.
- Partitioning: Add a salt mixture (e.g., 4g MgSO4, 1g NaCl), immediately shake for 1 minute, and centrifuge at 4000 rpm for 5 minutes.
- Cleanup: Transfer the supernatant to a dispersive Solid-Phase Extraction (d-SPE) tube containing primary secondary amine (PSA) and MgSO4. Vortex and centrifuge.
- LC-MS/MS Analysis:
- Column: C18 reversed-phase (e.g., 2.1 x 100 mm, 1.8 µm)
- Mobile Phase A: Water with 5mM ammonium formate/0.1% formic acid
- Mobile Phase B: Methanol with 5mM ammonium formate/0.1% formic acid
- Gradient: 5% B to 95% B over 10-15 minutes.
- Ionization: Electrospray Ionization (ESI) in positive or negative mode.
- Data Acquisition: Operate in MRM mode. For each pesticide, optimize the mass spectrometer to select the precursor ion and two characteristic product ions.
Data Analysis: Quantify pesticides against a calibration curve prepared with internal standards. Confirm identity based on retention time and the ratio of the two MRM transitions.
Protocol for GC-MS/MS Analysis of Pesticide Residues
Principle: Pesticides are extracted and, if necessary, derivatized to increase volatility. They are then separated by gas chromatography and detected by mass spectrometry, often using electron ionization and MRM for confirmation [18].
Materials & Reagents:
- Fruit peel samples
- GC-MS grade solvents: Acetonitrile, Ethyl Acetate
- Derivatization reagents (if needed): e.g., MSTFA for silylation
- QuEChERS extraction kits
- Internal Standards: Deuterated or other stable isotope-labeled standards
- Equipment: GC-MS/MS system, analytical balance, centrifuge, vortex mixer, derivatization heater
Procedure:
- Sample Preparation & Extraction: Follow steps 1-4 of the LC-MS/MS protocol.
- Derivatization (if required): For pesticides with active hydrogens (e.g., containing -OH, -COOH groups), evaporate an aliquot of the cleaned extract to dryness under a gentle nitrogen stream. Reconstitute in pyridine and add a derivatizing agent like MSTFA. Heat at 60°C for 20-30 minutes.
- GC-MS/MS Analysis:
- Column: Mid-polarity fused silica capillary column (e.g., 5% phenyl polysiloxane, 30m x 0.25mm i.d., 0.25µm film thickness)
- Carrier Gas: Helium or Hydrogen at 1.0 mL/min constant flow.
- Inlet Temperature: 250°C, operated in splitless mode.
- Oven Program: 60°C (hold 1 min), ramp to 300°C at 15-20°C/min, hold for 5-10 min.
- Transfer Line Temperature: 280°C.
- Ionization: Electron Ionization (EI) at 70 eV.
- Data Acquisition: Operate in MRM mode or use Selected Ion Monitoring (SIM) for higher sensitivity.
Data Analysis: Quantify by comparing the peak area of the target analyte to the internal standard. Confirm identity by comparing the sample spectrum to a certified library spectrum and verifying retention time.
The workflow for selecting and executing the appropriate confirmatory method is summarized below.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful analysis requires carefully selected reagents and materials. The following table lists key solutions used in the featured experiments.
Table 2: Essential Research Reagent Solutions for LC/GC-MS/MS Analysis of Pesticide Residues
| Reagent/Material | Function/Purpose | Example in Protocol |
|---|---|---|
| Stable Isotope-Labeled Internal Standards (e.g., ¹³C, ²H) | Correct for analyte loss during preparation and instrument variability; essential for high-accuracy quantification [73]. | Added before extraction in both LC-MS/MS and GC-MS/MS protocols. |
| QuEChERS Kits | Provide a standardized, efficient method for Quick, Easy, Cheap, Effective, Rugged, and Safe sample extraction and cleanup from complex food matrices [18]. | Used for the initial extraction and d-SPE cleanup steps. |
| Derivatization Reagents (e.g., MSTFA, BSTFA) | Chemically modify non-volatile pesticides to create volatile, thermally stable derivatives amenable to GC-MS analysis [70]. | Used in the GC-MS protocol for pesticides with -OH or -COOH groups. |
| LC-MS Grade Solvents & Additives | Provide high-purity mobile phase components to minimize background noise and ion suppression in the mass spectrometer. | Acetonitrile, Methanol, Water with 0.1% Formic Acid. |
| GC-MS Inlet Liners | Provide a deactivated surface for sample vaporization; a critical consumable for maintaining peak shape and sensitivity. | Replaced regularly to prevent performance degradation. |
LC-MS/MS and GC-MS/MS are powerful, complementary techniques that form the bedrock of modern analytical chemistry. LC-MS/MS excels in the analysis of polar, thermally labile, and high-molecular-weight compounds, while GC-MS/MS is unmatched for volatile and semi-volatile analytes. The choice between them is not a matter of superiority but of appropriateness for the analytical question at hand. In the context of validating a novel electrochemical biosensor for fruit pesticides, a judicious selection—or combination—of these techniques is imperative to provide the robust, reference-quality data needed to confirm the biosensor's accuracy and reliability. Understanding their respective performance trade-offs in sensitivity, scope, sample preparation, and cost is fundamental to effective analytical method design.
Within the broader research on developing electrochemical biosensor protocols for detecting pesticide residues in fruits, the validation of these sensors using real and spiked samples represents a critical step from laboratory innovation to practical application. Analytical techniques such as High-Performance Liquid Chromatography (HPLC) or Mass Spectrometry (MS) have traditionally been used for this purpose, but they involve complex procedures, high costs, long analysis times, and require complex sample pretreatment [51] [32]. Electrochemical biosensors have attracted considerable attention as alternatives due to their simplicity, rapidity, cost-effectiveness, portability, and appropriateness for real-time and on-site analysis [51] [74].
However, the performance of biosensors is greatly affected by the sample matrix itself, which can impact the accuracy and sensitivity of the measurements [6]. Therefore, to acquire reliable and accurate measurements, matrix effects and their influence on sensor performance must be thoroughly investigated. This application note details the experimental protocols and assessment criteria for validating electrochemical biosensors using spiked and real fruit samples, ensuring data reliability for researchers and scientists in the field of food safety and analytical chemistry.
Experimental Protocols
Biosensor Fabrication and Principle of Operation
The operational principle of many electrochemical biosensors for pesticide detection is based on enzyme inhibition. Acetylcholinesterase (AChE) is a commonly used enzyme whose activity is inhibited by organophosphate and carbamate pesticides [6].
Protocol: Fabrication of an AChE-based Electrochemical Biosensor
- Electrode Preparation: Use a three-electrode system, typically with a Gold (Au) or Glassy Carbon (GC) working electrode, a Platinum counter electrode, and an Ag/AgCl reference electrode. Screen-printed electrodes (SPEs) are cost-effective and attractive for portable devices [51].
- Surface Modification: Modify the working electrode surface with nanomaterials to enhance conductivity and surface area. For instance, conductive carbon black (CB) or carboxylated multi-walled carbon nanotubes (c-MWCNT) can be used [6].
- Enzyme Immobilization: Immobilize the biorecognition element (e.g., AChE enzyme) onto the modified working electrode. This can be achieved by:
- Preparing a dispersion of the nanomaterial (e.g., 2 mg/mL CB) in a dilute chitosan (CS) solution [6].
- Mixing this dispersion with the AChE enzyme solution.
- Depositing a precise volume (e.g., 5 μL) of the final mixture onto the working electrode surface and allowing it to dry.
- Cross-linking the enzyme with a glutaraldehyde solution to ensure stable immobilization.
- Storage: Store the fabricated biosensors at 4°C when not in use.
Sample Preparation and Extraction
A modified QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) method is widely applicable for preparing fruit samples for pesticide residue analysis [75] [76].
Protocol: QuEChERS-based Sample Preparation for Fruit Matrices
- Homogenization: Take a representative sample of the fruit (e.g., mandarin orange, grapefruit, apple) and homogenize it thoroughly.
- Extraction: Weigh 10.0 g ± 0.1 g of the homogenized sample into a 50 mL centrifuge tube. Add 10 mL of an extraction solvent (e.g., acetonitrile, or 0.1% formic acid in acetonitrile). Vortex vigorously for 1-2 minutes.
- Partitioning: Add a pre-made salts packet (e.g., containing 1.5 g sodium acetate and 4 g magnesium sulfate for the AOAC method, or 1 g sodium chloride, 4 g magnesium sulfate, 1 g sodium citrate, and 0.5 g disodium citrate sesquihydrate for the EN method) to induce liquid-liquid partitioning. Shake immediately and vigorously for 1 minute.
- Centrifugation: Centrifuge the tube at >4000 rpm for 5 minutes to separate the organic layer.
- Clean-up: Transfer an aliquot (e.g., 1 mL) of the upper acetonitrile layer into a dispersive Solid-Phase Extraction (d-SPE) tube containing cleanup sorbents. The choice of d-SPE sorbents (e.g., 150 mg MgSO₄ and 25 mg PSA; with or without C18 or GCB for specific pigments) should be optimized for the fruit matrix [75]. Vortex for 30-60 seconds and centrifuge.
- Preparation for Analysis: The cleaned extract is now ready for analysis. For electrochemical biosensing, further dilution with a suitable buffer (e.g., phosphate buffer saline, PBS) may be required to mitigate the inhibitory effect of the organic solvent on the enzyme [6].
Validation Procedure: Recovery and Accuracy Assessment
The core of the validation involves assessing the biosensor's performance by spiking blank fruit samples with known concentrations of target pesticides and calculating the recovery rate.
Protocol: Recovery Test for Method Validation
- Identification of a Blank Matrix: Source and analytically confirm a fruit sample that does not contain the target pesticide(s). This serves as the blank matrix.
- Sample Fortification (Spiking): Fortify the blank matrix with known concentrations of the pesticide standard solution at least five replicates per concentration level. Common spiking levels are 0.010 mg/kg (the Maximum Residue Limit, MRL, in many positive list systems), 0.080 mg/kg, and a higher level relevant to the sensor's range [75] [76]. Spiking can be done before the extraction process (to evaluate the entire method) or into the final extract (to evaluate the detection step alone).
- Analysis: Analyze the spiked samples using the fabricated electrochemical biosensor and the prepared sample extraction protocol. Common electrochemical techniques include Chronoamperometry (CA), Differential Pulse Voltammetry (DPV), Square-Wave Voltammetry (SWV), and Electrochemical Impedance Spectroscopy (EIS) [51].
- Calculation of Recovery: Calculate the percentage recovery for each spiked sample using the formula:
- Recovery (%) = (Measured Concentration / Spiked Concentration) × 100
- Accuracy Assessment: The mean recovery and the Relative Standard Deviation (RSD) of the replicates are calculated. According to validation guidelines like SANTE/12682/2019, acceptable recovery ranges are typically 70-120% with an RSD of ≤20% for the analytes at each spiking level [75] [76].
Addressing Matrix Effects
Matrix effects are a critical challenge, where components co-extracted from the fruit can interfere with the biosensor's signal, leading to inaccurate quantification [6].
Protocol: Evaluation and Mitigation of Matrix Effects
- Assessment: Compare the calibration curve prepared in the blank fruit matrix extract with the calibration curve prepared in a pure buffer solution. A significant difference in the slope indicates a matrix effect.
- Mitigation Strategies:
- Matrix-Matched Calibration: Prepare the calibration standards in the blank matrix extract. This is the most common and effective approach to compensate for matrix effects [75].
- Sample Dilution: Dilute the final sample extract with buffer to reduce the concentration of interfering compounds, provided the method's sensitivity allows it [75].
- Optimized Clean-up: Re-optimize the d-SPE clean-up step (e.g., type and amount of sorbents) to remove more specific interferents from the complex fruit matrix [76].
Data Presentation
The following tables consolidate quantitative recovery and accuracy data from research employing rigorous validation protocols for pesticide detection in fruits.
Table 1: Summary of Recovery Data for Pesticide Residues in Citrus Fruits using LC-MS/MS (Reference Method)
| Fruit Matrix | Number of Pesticides Analyzed | Spiking Level (mg/kg) | Average Recovery (%) | Relative Standard Deviation (RSD, %) | Citation |
|---|---|---|---|---|---|
| Mandarin Orange | 287 | 0.01 (PLS level) | 70 - 120 | ≤ 20 | [75] |
| Grapefruit | 287 | 0.01 (PLS level) | 70 - 120 | ≤ 20 | [75] |
| Orange | ~220 | 0.010 | Satisfied SANTE criteria | Satisfied SANTE criteria | [76] |
Table 2: Performance of Electrochemical Biosensors for Pesticide Detection
| Biosensor Type / Target | Sample Matrix | Linear Range | Limit of Detection (LOD) | Recovery in Real Samples (%) | Citation |
|---|---|---|---|---|---|
| AChE-based / Carbofuran | Vegetable Oils | Not specified | Not specified | Highly reproducible (with matrix-matched calibration) | [6] |
| c-MWCNT/Fe₃O₄/AChE / Malathion, Chlorpyrifos | Not specified | Not specified | 0.1 nM | Stable for 2 months | [32] |
| Aptamer-based / Carbendazim | Not specified | 0.003 - 10.0 μM | 1.0 nM | Not specified | [32] |
| MIP-based / Amino Acids, Vitamins | Sweat | Not specified | Trace levels | Correlated with serum levels | [77] |
The Scientist's Toolkit
Table 3: Essential Research Reagents and Materials
| Item | Function / Application | Examples / Specifications |
|---|---|---|
| Acetylcholinesterase (AChE) | Biorecognition element; its inhibition is measured to quantify pesticides. | From electric eel; immobilized on electrode surface. |
| Screen-Printed Electrodes (SPEs) | Portable, cost-effective sensing platform; working, counter, and reference electrode integrated. | Gold, Carbon, or Indium Tin Oxide (ITO) working electrodes. |
| QuEChERS Kits | Standardized sample preparation for extraction and clean-up of pesticides from complex fruit matrices. | AOAC 2007.01 or EN 15662 method kits; d-SPE kits with PSA, C18, GCB. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic, antibody-like recognition elements; offer high stability and selectivity for target analytes. | Used in wearable sensors for metabolites and nutrients [77]. |
| Aptamers | Single-stranded DNA/RNA oligonucleotides as recognition elements; high affinity and specificity for targets. | Selected for pesticides like carbendazim; used in electrochemical aptasensors [51] [32]. |
| Nanomaterials | Enhance electrode conductivity, surface area, and catalytic activity, improving sensor sensitivity. | Gold Nanoparticles (AuNPs), Carbon Nanotubes (c-MWCNT), Metal-Organic Frameworks (MOFs), Magnetic Nanoparticles (Fe₃O₄) [32]. |
Workflow and Signaling Diagrams
Experimental Workflow for Biosensor Validation
The following diagram illustrates the comprehensive workflow from sample preparation to validation assessment.
Signaling Principle of an AChE-based Biosensor
This diagram outlines the fundamental signaling mechanism behind acetylcholinesterase-based biosensors for pesticide detection.
For researchers developing electrochemical biosensors for fruit pesticide residues, a deep understanding of the regulatory landscapes governed by the European Food Safety Authority (EFSA) and the U.S. Environmental Protection Agency (EPA) is paramount. While the U.S. Food and Drug Administration (FDA) enforces these standards in the US, the EPA is responsible for setting the pesticide tolerances (the U.S. equivalent of Maximum Residue Limits or MRLs) [78] [79]. These legal standards represent the highest permissible level of pesticide residue in or on food, ensuring consumer safety when pesticides are applied according to Good Agricultural Practices (GAP) [80]. The development of analytical detection methods, including emerging biosensor technologies, must align with the stringent requirements and evolving updates of these global frameworks to ensure real-world applicability and compliance.
Recent monitoring data underscores the critical importance of reliable detection methods. In the European Union, random sampling of commonly consumed foods revealed that 96.3% of analyzed samples fell within legally permitted MRLs, with a subset from the coordinated control program showing an even higher compliance rate of 98.4% [81]. Similarly, for the 2023 monitoring cycle, 99% of random samples were compliant with EU legislation [82]. These figures highlight both the generally high rate of regulatory adherence and the continued need for precise detection capabilities to identify the approximately 1-3% of samples that exceed legal limits, ensuring food safety and regulatory compliance.
Current Regulatory Status and Monitoring Data
EFSA MRL Compliance and Monitoring Findings
The European Food Safety Authority employs a comprehensive monitoring program that combines random sampling under the EU-coordinated control programme (EU MACP) with targeted risk-based sampling through the Multiannual National Control Programme (MANCP). The most recent data indicates consistent compliance with MRL regulations across the European market.
Table 1: EFSA Pesticide Residue Monitoring Results (2022-2023)
| Program | Sampling Year | Total Samples | Within MRLs | Exceeded MRLs | Key Commodities Sampled |
|---|---|---|---|---|---|
| EU MACP (Random) | 2022 | 110,829 | 96.3% | 1.6% (1,192 samples) | Apples, strawberries, peaches, wine, lettuce, tomatoes, spinach [81] |
| EU MACP (Random) | 2023 | 13,246 | 99% | 2% (1% non-compliant after uncertainty) | Carrots, cauliflowers, kiwifruits, oranges, pears, potatoes [82] |
| MANCP (Targeted) | 2023 | 132,793 | 98% | 3.7% (2% non-compliant) | Risk-based sampling of various commodities [82] |
EFSA's dietary risk assessment, which incorporates these monitoring results, consistently concludes that there is a low risk to consumer health from estimated exposure to pesticide residues in the tested foods [81] [82]. The authority recommends continued monitoring of pesticide-crop combinations that frequently lead to non-compliances, particularly for imported products.
US EPA Tolerance Setting and FDA Enforcement
In the United States, the Environmental Protection Agency (EPA) establishes pesticide tolerances under the Federal Food, Drug, and Cosmetic Act (FFDCA) [79]. The FDA and USDA then enforce these tolerances through monitoring and surveillance programs [78]. The EPA's tolerance setting process requires a comprehensive safety finding of "reasonable certainty of no harm" based on extensive scientific data including pesticide toxicity, application patterns, residue persistence, and aggregate exposure from all sources [79].
Table 2: Recent MRL Updates in Key Export Markets (2025)
| Market | Pesticide (Example Trade Name) | MRL Change | Previous MRL | New MRL |
|---|---|---|---|---|
| Canada | Sulfoxaflor (Transform) | Raised/Harmonized | 0.1 ppm | 2.0 ppm [83] |
| European Union | Mefentrifluconazole (Cevya) | Raised/Harmonized | 0.01 ppm | 5.0 ppm [83] |
| Japan | Iprodione | Expired | 15 ppm | 0.01 ppm [83] |
| Philippines | Zeta-Cypermethrin (Mustang) | Established | No MRL | 1.5 ppm [83] |
| United States | Flonicamid (Beleaf) | Tolerance Set | No U.S. tolerance | 1.5 ppm [83] |
International MRLs are dynamic, with frequent updates that researchers and exporters must monitor closely. The USDA Maximum Residue Limits (MRL) Database provides a centralized resource for checking current tolerances across multiple countries, though users are advised to verify information with knowledgeable parties in the target market prior to sale or shipment [84].
Experimental Protocols for MRL Compliance Validation
Sample Preparation and Extraction Workflow
Proper sample preparation is critical for accurate pesticide residue detection. The following protocol, adapted from QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) methodology, is widely used in regulatory testing and should be optimized for biosensor applications.
Materials Required:
- Homogenizer or high-speed blender
- Analytical balance (±0.0001 g precision)
- Centrifuge capable of 4000-5000 rpm
- Vortex mixer
- QuEChERS extraction kits (containing MgSO4, NaCl)
- Dispersive SPE cleanup kits (containing PSA, C18, MgSO4)
- Calibrated pipettes and disposable tips
- Solvents: HPLC-grade acetonitrile, methanol, acetone
- Internal standards solution (e.g., triphenyl phosphate)
Procedure:
- Sample Homogenization: Representative fruit samples (minimum 1 kg) are thoroughly homogenized using a high-speed blender to ensure a uniform matrix.
- Subsampling: Accurately weigh 15.0 ± 0.1 g of homogenized sample into a 50 mL centrifuge tube.
- Extraction: Add 15 mL of acetonitrile (1% acetic acid) and internal standard solution. Vortex vigorously for 1 minute.
- Salting Out: Add extraction salts (6 g MgSO4, 1.5 g NaCl), immediately shake for 1 minute, and centrifuge at 4000 rpm for 5 minutes.
- Cleanup: Transfer 8 mL of supernatant to a d-SPE tube (containing 400 mg PSA, 400 mg C18, 1200 mg MgSO4). Vortex for 1 minute and centrifuge at 4000 rpm for 5 minutes.
- Preparation for Analysis: Transfer the purified extract to a autosampler vial for analysis. For electrochemical biosensing, further dilution in appropriate buffer may be necessary to ensure compatibility with the sensing platform.
This sample preparation method effectively reduces matrix interference, a significant challenge in electrochemical detection, particularly for complex fruit matrices [5].
Electrochemical Biosensor Calibration and Validation Protocol
Calibration against reference standards and validation according to regulatory guidelines is essential for method acceptance. This protocol outlines the procedure for establishing a reliable calibration curve and validating biosensor performance for pesticide detection.
Materials Required:
- Certified pesticide reference standards (purity >95%)
- Appropriate solvent for stock solution preparation
- Supporting electrolyte (e.g., phosphate buffer, acetate buffer)
- Electrochemical cell or microfluidic chip with integrated electrodes
- Potentiostat/galvanostat with impedance capability
- Standard solutions for comparison (if using chromatographic validation)
Procedure:
- Stock Solution Preparation: Accurately weigh 10 mg of certified pesticide standard and dissolve in 10 mL of appropriate solvent to create 1000 ppm stock solution. Store at -20°C.
- Calibration Standards: Prepare serial dilutions in electrolyte buffer to cover the expected concentration range (e.g., 0.1 ppb to 1000 ppb). Include concentrations spanning the target MRL.
- Biosensor Measurement:
- Immerse the working electrode in electrolyte buffer and record baseline.
- Add calibrated standard, incubate with stirring for fixed time (e.g., 5-15 minutes).
- Perform electrochemical measurement (e.g., DPV, EIS, amperometry).
- Rinse electrode between measurements with appropriate regeneration buffer.
- Calibration Curve: Plot sensor response (current, impedance change) versus logarithm of concentration. Determine linear range, limit of detection (LOD), and limit of quantification (LOQ).
- Validation:
- Accuracy: Perform recovery studies using spiked samples at 0.5x, 1x, and 2x MRL concentrations.
- Precision: Assess repeatability (intra-day) and reproducibility (inter-day) with relative standard deviation (RSD).
- Specificity: Test against structurally similar compounds and common fruit matrix interferents.
For regulatory acceptance, the developed biosensor method should demonstrate performance characteristics comparable to established reference methods like GC-MS or LC-MS [7], with particular attention to achieving LODs sufficiently below the target MRLs to ensure reliable compliance monitoring.
Research Reagent Solutions for Electrochemical Biosensing
Table 3: Essential Research Reagents for Pesticide Residue Biosensor Development
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Enzyme Probes (AChE, ChO) | Biospecific recognition element for organophosphates/carbamates | Inhibition-based detection; requires stability optimization [7] |
| Antibody Probes | High-affinity molecular recognition for specific pesticides | Immunosensor development; requires careful conjugate design [5] |
| Aptamer Sequences | Synthetic oligonucleotide recognition elements | Aptasensors; offer stability and design flexibility [5] |
| Nanomaterial Modifiers (Graphene, CNTs, Metal NPs) | Electrode surface modification to enhance sensitivity | Increase active surface area and electron transfer kinetics [7] |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymer with tailored recognition cavities | Biomimetic sensors; offer excellent stability [5] |
| Electrochemical Redox Probes ([Fe(CN)₆]³⁻/⁴⁻) | Electron transfer mediator for signal generation | Impedimetric and voltammetric detection; concentration optimization required [7] |
| Enzyme Substrates (Acetylthiocholine) | Substrate for enzymatic generation of electroactive product | Inhibition-based assays; concentration affects sensitivity [7] |
Signaling Pathways and Experimental Workflows
Electrochemical Biosensor Operation Workflow
The following diagram illustrates the complete experimental workflow from sample preparation to electrochemical detection and data analysis, highlighting critical steps where regulatory considerations impact protocol design.
Regulatory Framework Integration Pathway
This diagram maps the critical interaction points between biosensor development and regulatory frameworks, emphasizing how MRL standards influence method validation and application.
The development of electrochemical biosensors for pesticide residue detection in fruits must be intrinsically linked to the dynamic regulatory frameworks established by EFSA, EPA, and other international bodies. The experimental protocols and reagent solutions outlined in this application note provide researchers with a foundation for developing detection methods that are not only analytically sensitive but also regulatory relevant. By aligning biosensor validation with established MRL compliance requirements and incorporating current monitoring data, researchers can bridge the gap between technological innovation and practical implementation in food safety systems. Future directions should focus on multiplexed detection capabilities to address the complex residue profiles encountered in real-world samples, miniaturization for field-deployable compliance screening, and enhanced data integration with regulatory databases to facilitate rapid decision-making.
Multiplexed Electrochemical Aptasensor for Simultaneous Pesticide Detection
The need to detect multiple pesticide residues simultaneously in fruit has become a critical challenge in food safety analysis. Traditional methods, such as gas chromatography (GC) or high-performance liquid chromatography (HPLC), are often limited to single-component detection, making the process time-consuming, labor-intensive, and costly for multi-residue screening [85] [86]. Multiplexed electrochemical biosensors address this limitation by enabling the parallel, quantitative detection of several analytes in a single measurement, significantly shortening analysis time, reducing costs, and achieving high-efficiency analysis [85] [87]. These sensors are particularly suited for on-site screening and provide a rapid, cost-effective, and portable alternative to conventional laboratory techniques [86] [51]. The core principle involves using specific biorecognition elements, such as aptamers, coupled with distinguishable electrochemical signal probes to generate independent signals for different target pesticides without mutual interference [85] [88].
Core Principles and Signaling Mechanisms
Achieving multiplexing requires the integration of specific biorecognition elements with distinguishable electrochemical probes. Aptamers, which are short, single-stranded DNA or RNA oligonucleotides, are ideal for this purpose due to their high stability, ease of chemical modification, and excellent specificity [85] [86]. They are selected to bind specifically to different target pesticides.
For signal generation and differentiation, electrochemical probes that produce signals at distinct, non-overlapping potentials are crucial. A prominent strategy utilizes different electroactive tags. For instance, in a sensor for malathion and chlorpyrifos, thionine (Thi) and ferrocene (Fc) were used as probes [85]. Thionine generates a signal at a lower potential, while ferrocene produces one at a higher potential, allowing for simultaneous and independent detection in the same solution [85]. Signal amplification is often achieved using nanomaterials. Mixed-valence metal-organic frameworks (MOFs), such as Ce(III, IV)-MOF, provide a high surface area for loading numerous probe molecules and possess intrinsic catalytic properties that can further enhance the electrochemical signal, thereby improving sensitivity [85].
The following diagram illustrates the general workflow and signaling mechanism of a multiplexed aptasensor.
Detailed Experimental Protocols
Protocol A: Ce-MOF-Based Aptasensor for Malathion and Chlorpyrifos
This protocol details the construction of a dual-analyte sensor using mixed-valence Ce-MOF for signal amplification [85].
3.1.1 Materials and Reagents
- Trimesic acid (TMA)
- Cerium(III) nitrate hexahydrate (Ce(NO₃)₃·6H₂O)
- Thionine (Thi) and Ferrocene (Fc): Electrochemical probes.
- Chlorpyrifos and Malathion aptamers (Apt1 & Apt2): Biorecognition elements.
- Complementary DNA (cDNA): Modified with a thiol or amino group for immobilization on the electrode.
- 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) / N-Hydroxysuccinimide (NHS): For activating carboxyl groups to form amide bonds with aptamers.
- Screen-printed gold or carbon electrodes: Transducer platform.
3.1.2 Step-by-Step Procedure
- Synthesis of Ce(III, IV)-MOF: Dissolve TMA (0.210 g) in 10 mL of a water-ethanol mixed solution (1:1 v/v). Add 10 mL of Ce(NO₃)₃·6H₂O solution (0.5 mM) to the mixture with stirring. Centrifuge the resulting mixture at 8500 rpm for 8 minutes, wash the precipitate with deionized water, and dry at 60°C to obtain the MOF material [85].
- Preparation of Signal Probes:
- Chlorpyrifos Probe: Load thionine and the chlorpyrifos aptamer (Apt1) onto the Ce(III, IV)-MOF.
- Malathion Probe: Load ferrocene and the malathion aptamer (Apt2) onto the Ce(III, IV)-MOF.
- Electrode Modification and Aptasensor Assembly:
- Immobilize the cDNA on the cleaned electrode surface.
- Hybridize the signal probes (from Step 2) with the immobilized cDNA on the electrode. This step fixes the probes to the electrode surface via base pairing.
- Measurement and Detection:
- Incubate the assembled aptasensor with the sample solution containing chlorpyrifos and malathion.
- The specific binding of targets to their aptamers causes the signal probes to be released from the electrode surface, leading to a change in the electrochemical signal.
- Record signals using Differential Pulse Voltammetry (DPV). The reduction of thionine (catalyzed by the Ce(III/IV) cycle) and ferrocene (catalyzed by ascorbic acid in solution) generates distinct current peaks at different potentials [85].
Protocol B: rGO-Based Aptasensor for Neonicotinoids
This protocol describes a multiplexed sensor for three neonicotinoid pesticides using reduced graphene oxide (rGO) to enhance sensitivity [88].
3.2.1 Materials and Reagents
- Graphene Oxide (GO) dispersion
- 1-Pyrenebutyric Acid (Py): A linker molecule for functionalizing rGO.
- Amine-labeled aptamers specific to imidacloprid, thiamethoxam, and clothianidin.
- EDC/NHS crosslinking chemistry.
- Screen-printed carbon electrodes (SPCEs)
3.2.2 Step-by-Step Procedure
- Electrode Modification with rGO:
- Drop-coat the GO dispersion onto the SPCE.
- Electrochemically reduce GO to rGO to improve conductivity.
- Functionalization and Aptamer Immobilization:
- Treat the rGO/SPCE with 1-pyrenebutyric acid. The pyrene group interacts strongly with the rGO surface via π-π stacking, exposing carboxylic acid groups.
- Activate the carboxyl groups using EDC/NHS.
- Covalently immobilize the amine-labeled aptamers for imidacloprid, thiamethoxam, and clothianidin onto the activated surface.
- Measurement and Detection:
- After incubating with the sample, measure the sensor response using Differential Pulse Voltammetry (DPV) in a solution containing
[Fe(CN)₆]³⁻/⁴⁻as a redox probe. - The binding of target pesticides to the aptamers alters the electron transfer kinetics of the redox probe at the electrode interface. This change is measured as a shift in current, which is proportional to the analyte concentration [88].
- After incubating with the sample, measure the sensor response using Differential Pulse Voltammetry (DPV) in a solution containing
Performance Data and Comparison
The quantitative performance of recently reported multiplexed electrochemical aptasensors is summarized in the table below.
Table 1: Performance Metrics of Multiplexed Electrochemical Aptasensors for Pesticides
| Target Pesticides | Sensor Platform | Linear Detection Range | Limit of Detection (LOD) | Detection Technique | Citation |
|---|---|---|---|---|---|
| Chlorpyrifos & Malathion | Ce(III, IV)-MOF Aptasensor | 1.0 μM ~ 0.1 pM | 0.038 pM (Chlorpyrifos)0.045 pM (Malathion) | DPV | [85] |
| Imidacloprid, Thiamethoxam & Clothianidin | Reduced Graphene Oxide (rGO) Aptasensor | 0.01 ng/mL to 100 ng/mL | Not specified (excellent sensitivity reported) | DPV | [88] |
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents and materials essential for developing multiplexed electrochemical aptasensors.
Table 2: Key Research Reagent Solutions for Multiplexed Aptasensor Development
| Reagent/Material | Function and Role in the Experiment |
|---|---|
| Screen-Printed Electrodes (SPEs) | Provide a cost-effective, disposable, and miniaturized platform for sensor fabrication, ideal for portability and on-site analysis [51]. |
| Specific Aptamers | Serve as the biorecognition element that binds specifically to target pesticide molecules. Their chemical stability and modifiability are crucial for multiplexing [85] [88]. |
| Electrochemical Probes (e.g., Thionine, Ferrocene) | Act as signal tags that generate distinguishable electrochemical signals (at different potentials) for each target analyte, enabling simultaneous detection [85]. |
| Signal Amplification Nanomaterials (e.g., Ce-MOF, rGO) | Enhance sensor sensitivity. MOFs offer high surface area and catalytic activity [85], while rGO provides excellent conductivity and a large surface for biomolecule immobilization [88]. |
| Crosslinkers (e.g., EDC/NHS) | Facilitate the covalent immobilization of biomolecules (like amine-labeled aptamers) onto functionalized electrode surfaces, ensuring stable sensor assembly [88]. |
Troubleshooting and Technical Notes
- Signal Overlap: If the DPV peaks for different probes overlap, optimize the types and concentrations of the electrochemical probes. Ensure their peak potentials are sufficiently separated (e.g., >100 mV).
- High Background Noise: Thoroughly wash the electrode after each immobilization and incubation step to remove unbound reagents. Verify the cleanliness of buffers and solutions.
- Low Sensitivity or Signal Response: Check the activity of the immobilized aptamers and the effectiveness of the nanomaterial coating on the electrode. Ensure proper assembly of the signal probes.
- Validation: Always validate the sensor's performance in real fruit samples (e.g., spiked recovery tests) and correlate results with a standard method like LC-MS/MS to ensure accuracy [88].
Conclusion
Electrochemical biosensors represent a paradigm shift in pesticide residue analysis, offering a powerful, decentralized alternative to traditional laboratory-bound methods. This synthesis of foundational knowledge, methodological protocols, optimization strategies, and validation frameworks underscores their potential for rapid, sensitive, and on-site monitoring, directly contributing to enhanced food safety. Future directions crucial for clinical and biomedical translation include the integration of AI-driven data analytics for improved pattern recognition, the development of fully integrated, user-friendly portable devices for field use, and the pursuit of multiplexed platforms for comprehensive contaminant profiling. The continued convergence of nanotechnology, materials science, and biotechnology is poised to further revolutionize this field, enabling precise, preventative public health protection.
References
- 1. Assessment of Organophosphate and Carbamate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3673447/]
- 2. Piezoelectric Biosensors for Organophosphate and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4264360/]
- 3. Detection of Organophosphorus, Pyrethroid, and ... [https://www.mdpi.com/2304-8158/14/14/2543]
- 4. Recent developments in monitoring of organophosphorus ... [https://www.sciencedirect.com/science/article/pii/S2666154325000808]
- 5. Pesticide residue detection techniques for increasing ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1694779/full]
- 6. AChE-based electrochemical biosensor for pesticide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9839810/]
- 7. Electrochemical detection of pesticides [https://www.sciencedirect.com/science/article/pii/S1388248124001826]
- 8. Molecularly Imprinted Polymer-Enhanced Electrochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12631419/]
- 9. What are the Main Sensor Methods for Quantifying Pesticides ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6680408/]
- 10. Organophosphorus pesticide detection directly on fruit peels [https://www.sciencedirect.com/science/article/pii/S0039914024014723]
- 11. Biomaterials and biosensing technologies in the detection ... [https://www.sciencedirect.com/science/article/abs/pii/S0010854525006800]
- 12. Research Advances in Nanosensor for Pesticide Detection in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12298915/]
- 13. Seeing Neurodegeneration in a New Light Using Genetically ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9861453/]
- 14. Research Progress of Biosensors in the Detection ... [https://www.mdpi.com/2079-6374/15/12/778]
- 15. About One Health [https://www.cdc.gov/one-health/about/index.html]
- 16. A One Health Approach to Address Foodborne Diseases in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11951973/]
- 17. Recent Advances in Nanomaterial-Based Biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9405907/]
- 18. Innovative Analytical Approaches for Food Pesticide Residue ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12452724/]
- 19. Current Advances in Aptasensors for Pesticide Detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC11930883/]
- 20. Electrochemical Biosensors - Sensor Principles and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3663003/]
- 21. Electrochemical biosensors: recommended definitions and ... [https://www.sciencedirect.com/science/article/pii/S0956566301001154]
- 22. Electrochemical biosensors: perspective on functional ... [https://biomaterialsres.biomedcentral.com/articles/10.1186/s40824-019-0181-y]
- 23. What are Electrochemical Biosensors? [https://www.news-medical.net/life-sciences/What-are-Electrochemical-Biosensors.aspx]
- 24. Electrochemical Biosensors - an overview [https://www.sciencedirect.com/topics/engineering/electrochemical-biosensors]
- 25. Electrochemical Sensors and Their Applications: A Review [https://www.mdpi.com/2227-9040/10/9/363]
- 26. Recent Advances in Electrochemical Biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9775431/]
- 27. Recent developments in non-enzymatic (bio)sensors for ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914021003180]
- 28. Advances in Enzyme-Based Biosensors for Pesticide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6022933/]
- 29. Internet-of-things-integrated molecularly imprinted polymer ... [https://www.sciencedirect.com/science/article/abs/pii/S0269749124007437]
- 30. Aptamer-based sensor for specific recognition of malathion ... [https://www.sciencedirect.com/science/article/abs/pii/S000326702200719X]
- 31. Sensitive enzyme-free electrochemical sensors for the ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993624002115]
- 32. Progress of rapid detection of pesticides in fruits and ... [https://www.frontiersin.org/journals/food-science-and-technology/articles/10.3389/frfst.2023.1253227/full]
- 33. Graphene, carbon nanotubes, zinc oxide and gold as elite ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566315002146]
- 34. Signal multi-amplified electrochemical biosensor for ... [https://pubmed.ncbi.nlm.nih.gov/32350619/]
- 35. Carbon Nanomaterial-Based Electrochemical Biosensors ... [https://www.mdpi.com/2079-6374/15/10/684]
- 36. Functional Nanomaterials Enhancing Electrochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10180161/]
- 37. Quantitative analysis of the response of an electrochemical ... [https://www.sciencedirect.com/science/article/abs/pii/S095656630400421X]
- 38. Surface immobilization strategies for the development of ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566323003822]
- 39. Surface-Functionalizing Strategies for Multiplexed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11677728/]
- 40. Development of label-free electrochemical OMP-DNA ... [https://www.nature.com/articles/s41598-024-63112-w]
- 41. Recent advances in surface functionalization techniques ... [https://pubs.rsc.org/en/content/articlelanding/2014/an/c3an01789c]
- 42. Electrochemical biosensors based on the 3D ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12376872/]
- 43. Critical overview on the application of sensors and biosensors ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7112812/]
- 44. Innovative sample preparation techniques in food analysis [https://www.sciencedirect.com/science/article/pii/S2772582025000440]
- 45. Electrochemical (Bio)Sensors for Pesticides Detection ... [https://www.mdpi.com/2079-6374/10/4/32]
- 46. A review of smart electrochemical devices for pesticide ... [https://www.sciencedirect.com/science/article/abs/pii/S0048969724035071]
- 47. Organophosphorus pesticide detection directly on fruit peels [https://pubmed.ncbi.nlm.nih.gov/39486304/]
- 48. Glove detects chemical residues on food in a few minutes [https://www.futurefarming.com/crop-solutions/glove-detects-chemical-residues-on-food-in-a-few-minutes/]
- 49. Development of the screen-printed electrodes: A mini ... [https://www.sciencedirect.com/science/article/pii/S2352186422003455]
- 50. Electrochemical Biosensors Driving Model Transformation ... [https://www.mdpi.com/2304-8158/14/15/2669]
- 51. Electrochemical (Bio)Sensors for Toxins, Foodborne ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12293878/]
- 52. Self-Calibrating Dual-Signal Cascade Amplification ... [https://www.sciencedirect.com/science/article/abs/pii/S0927775725028985]
- 53. Acetylcholine triggered enzymatic cascade reaction based ... [https://www.sciencedirect.com/science/article/pii/S0003267024002654]
- 54. Organic-Dominated Nanozymes for Pesticide Detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC12532300/]
- 55. A Multi-Enzyme Cascade Response for the Colorimetric ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10486685/]
- 56. Electrical modeling of an electrochemical sensor operating ... [https://www.sciencedirect.com/science/article/abs/pii/S0013468624009150]
- 57. In Vivo Plant Bio-Electrochemical Sensor Using Redox ... [https://www.mdpi.com/2079-6374/13/2/219]
- 58. Surface-enhanced Raman spectroscopy biosensor ... [https://link.springer.com/article/10.1007/s44462-025-00031-7]
- 59. Review of pesticide residue analysis in fruits and ... [https://www.sciencedirect.com/science/article/abs/pii/S0963996920301666]
- 60. Fundamentals of bio-electrochemical sensing [https://www.sciencedirect.com/science/article/pii/S266682112300073X]
- 61. Determination of stability characteristics for electrochemical ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914017304253]
- 62. Improving the Stability and Sensing of Electrochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3956047/]
- 63. , Artificial Intelligence Biosensors , Biosensors Results and... false [https://pmc.ncbi.nlm.nih.gov/articles/PMC12025796/]
- 64. Cascade-responsive microsystems with multi-phase electron transfer... [https://www.sciopen.com/article/10.26599/NR.2025.94907753]
- 65. Biosensors in food processing - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC3671056/]
- 66. Biosensors Metrics Glossary of Definitions - Zimmer & Peacock [https://www.zimmerpeacocktech.com/knowledge-base/faq/biosensors-glossary/]
- 67. Determination of 301 pesticide residues in tropical fruits ... [https://www.sciencedirect.com/science/article/abs/pii/S0956713522007691]
- 68. Determination and dietary risk assessment of 284 pesticide ... [https://www.nature.com/articles/s41598-021-89204-5]
- 69. Quantitative determination and removal of pesticide ... [https://pubmed.ncbi.nlm.nih.gov/36609771/]
- 70. LCMS vs. GCMS: When to Choose Each for Optimal ... [https://emerypharma.com/blog/lcms-vs-gcms-when-to-choose-each-for-optimal-results-in-your-analytical-chemistry/]
- 71. The Difference Between GC/MS and LC/MS Systems [https://conquerscientific.com/the-difference-between-gc-ms-and-lc-ms-systems/]
- 72. Latest News on Mass Specs – February 2025 [https://www.phenomenex.com/news/news/2025/02/mass-specs?srsltid=AfmBOoqcHz3cP3CxQ-aE5NHakuIoASlbtGnJcPSs3qAe5Jw72wzwmZji]
- 73. Perspectives of Quantitative GC-MS, LC-MS, and ICP-MS in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11641993/]
- 74. Electrochemical biosensors: The beacon for food safety ... [https://www.sciencedirect.com/science/article/pii/S0308814625005357]
- 75. Validation of a Multi-Residue Analysis Method for 287 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9657228/]
- 76. Development and Validation of Pesticide Residues ... [https://www.mdpi.com/2077-0472/12/11/1936]
- 77. A wearable electrochemical biosensor for the monitoring of ... [https://www.nature.com/articles/s41551-022-00916-z]
- 78. Chemical Contaminants & Pesticides [https://www.fda.gov/food/chemical-contaminants-pesticides]
- 79. Setting Tolerances for Pesticide Residues in Foods [https://www.epa.gov/pesticide-tolerances/setting-tolerances-pesticide-residues-foods]
- 80. Maximum Residue Levels - Language selection | Food Safety [https://food.ec.europa.eu/plants/pesticides/maximum-residue-levels_en]
- 81. Pesticide residues in food: latest figures released - EFSA [https://www.efsa.europa.eu/en/news/pesticide-residues-food-latest-figures-released]
- 82. Pesticides residues in food: what's the situation in the EU? [https://www.efsa.europa.eu/en/news/pesticides-residues-food-whats-situation-eu]
- 83. 2025 Blueberry MRL Memo and Guidance Now Available [https://calfruitandveg.com/2025/04/15/2025-blueberry-mrl-memo-and-guidance-now-available/]
- 84. Maximum Residue Limits (MRL) Database [https://www.fas.usda.gov/maximum-residue-limits-mrl-database]
- 85. Multiplexed electrochemical aptasensor based on mixed ... [https://www.sciencedirect.com/science/article/abs/pii/S0003267022009357]
- 86. Recent advances in rapid detection techniques for pesticide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10584040/]
- 87. Multimodal Fusion-Driven Pesticide Residue Detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC12430378/]
- 88. Reduced graphene oxide-based electrochemical ... [https://www.nature.com/articles/s41598-025-94313-6]