Electrochemical Biosensors in Agriculture: A Comprehensive Review of Principles, Applications, and Future Directions for Smart Farming
This article provides a comprehensive overview of the transformative role of electrochemical biosensors in modern agriculture.
Electrochemical Biosensors in Agriculture: A Comprehensive Review of Principles, Applications, and Future Directions for Smart Farming
Abstract
This article provides a comprehensive overview of the transformative role of electrochemical biosensors in modern agriculture. It explores the foundational principles of these analytical devices, which combine a biological recognition element with an electrochemical transducer to convert a biological response into a quantifiable signal. The review details the diverse methodologies and specific applications in precision agriculture, including the real-time monitoring of plant health, early detection of devastating pathogens in key crops like oilseed rape and soybean, and analysis of soil and environmental conditions. It critically examines the technical challenges and optimization strategies for field deployment, such as overcoming matrix interference and improving sensor stability. Furthermore, the article offers a comparative analysis with traditional analytical methods, highlighting the superior portability, cost-effectiveness, and rapid response of biosensors. Finally, it discusses the integration of these sensors with emerging technologies like the Internet of Things (IoT), artificial intelligence (AI), and smartphone-based platforms, outlining a future roadmap for sustainable crop management and enhanced global food security.
The Fundamentals of Electrochemical Biosensors: Principles, Components, and Transduction Mechanisms
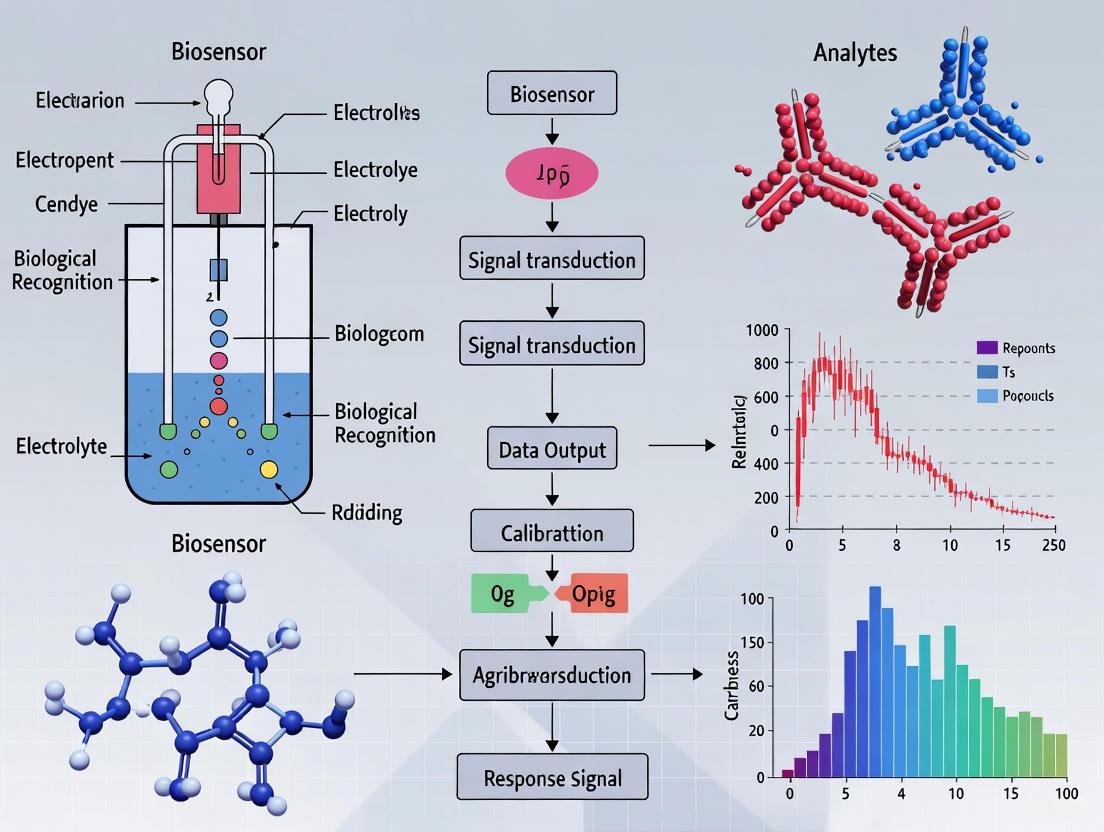
Electrochemical biosensors represent a powerful class of analytical devices that combine the specificity of biological recognition with the sensitivity of electrochemical transduction. These tools are transforming fields ranging from clinical diagnostics to environmental monitoring and, critically, agricultural research [1] [2]. For scientists engaged in agriculture, these biosensors offer the potential for real-time, on-site detection of pathogens, toxins, and stress biomarkers in crops and soil, enabling precision agriculture and early intervention strategies [3] [4]. The performance of any electrochemical biosensor hinges on the seamless integration of its core components: the bioreceptor, which provides molecular recognition, and the transducer, which converts the biological event into a quantifiable electrical signal [5] [6]. This whitepaper provides an in-depth technical guide to these fundamental elements, detailing their principles, configurations, and experimental implementation within an agricultural research context.
Core Components of an Electrochemical Biosensor
A typical electrochemical biosensor is an integrated device composed of five main elements: the bioreceptor, the interface, the transducer, detector electronics, and data output software [5] [6]. The central process involves the selective binding of the target analyte (e.g., a pathogen DNA sequence or a protein biomarker) by the bioreceptor immobilized on a sensor surface. This biological interaction produces a physicochemical change at the interface, which the transducer converts into an electrical signal. The detector circuit processes this signal, and software finally translates it into a meaningful physical parameter for the user [5]. The following diagram illustrates this workflow and the relationship between these core components.
The Bioreceptor: Engine of Molecular Recognition
The bioreceptor is the molecular recognition element of a biosensor, responsible for its high selectivity and specificity. It is a biological or biomimetic entity immobilized on the sensor surface that selectively binds to the target analyte [1] [5]. The choice of bioreceptor is dictated by the application and determines key sensor characteristics like stability, reproducibility, and susceptibility to interferences.
Common types of bioreceptors used in agricultural biosensing include:
- Enzymes: These biocatalysts recognize substrates and generate electroactive products (e.g., H₂O₂, O₂) whose concentration can be measured. They are widely used for detecting pesticides, which often act as enzyme inhibitors [5] [2].
- Antibodies: These immunoproteins form highly specific "lock-and-key" complexes with antigens (e.g., whole bacterial cells, viral proteins). Biosensors using antibodies are termed immunosensors and are central to pathogen detection [5] [7].
- Nucleic Acids (DNA/RNA): Single-stranded DNA or RNA probes hybridize with complementary target sequences, allowing for the detection of specific pathogen DNA (e.g., from Sclerotinia sclerotiorum) with high precision [1] [3].
- Whole Cells: Microorganisms (e.g., bacteria, yeast) or plant cells can serve as living bioreceptors, often engineered to produce an electrical signal in response to specific metabolic changes or environmental contaminants [5] [8].
- Aptamers: These are short, single-stranded oligonucleotides (DNA or RNA) selected in vitro for high-affinity binding to a target, from small molecules to whole cells. They are known as "chemical antibodies" and offer advantages in stability and synthesis [1] [3].
Table 1: Common Bioreceptors in Agricultural Electrochemical Biosensors
| Bioreceptor | Recognition Principle | Key Advantages | Agricultural Application Examples |
|---|---|---|---|
| Enzymes | Catalytic substrate conversion | High turnover = signal amplification | Detection of organophosphate pesticides [5] |
| Antibodies | Affinity-based antigen binding | High specificity and affinity | Detection of E. coli O157:H7, Salmonella [7] [9] |
| Nucleic Acids | Base-pair hybridization | High specificity, stable receptors | Detection of fungal pathogen DNA [3] |
| Aptamers | 3D structure-based affinity | High stability, synthetic production | Detection of mycotoxins [1] |
| Whole Cells | Metabolic or stress response | Functional, multi-parameter response | Engineered cell sensors for bioactive compounds [8] |
The Transducer: Principle of Electrochemical Signal Conversion
The transducer is the component that converts the biological recognition event into a measurable electrical signal. In electrochemical biosensors, this occurs via electrodes that are in contact with the analytical sample [5] [2]. The design of the electrode and the electrochemical technique applied are critical for sensitivity, detection limits, and suitability for field use.
The most common electrochemical transduction techniques are:
- Amperometry: Measures the current generated by the electrochemical oxidation or reduction of an electroactive species at a constant applied potential. The current is directly proportional to the concentration of the species [5] [6]. It is widely used in enzyme-based biosensors.
- Potentiometry: Measures the potential difference (voltage) between a working electrode and a reference electrode when little to no current flows between them. Ion-selective electrodes (e.g., pH electrodes) are a common example [5] [6].
- Impedance Spectroscopy (EIS): Measures the impedance (resistance to current flow) of the electrode interface. The binding of a target analyte to the bioreceptor often alters the interfacial impedance, allowing for label-free detection [5].
- Voltammetry: Measures the current while the potential between the electrodes is swept and the redox behavior of electroactive species is analyzed. Techniques like Differential Pulse Voltammetry (DPV) offer high sensitivity [5] [9].
- Conductometry: Measures the change in the electrical conductivity of a solution resulting from a biological reaction [5] [6].
Table 2: Core Electrochemical Transduction Techniques
| Technique | Measured Quantity | Principle | Advantages |
|---|---|---|---|
| Amperometry | Current | Redox reaction of electroactive species | High sensitivity, well-established |
| Potentiometry | Potential | Ion activity at electrode surface | Wide detection range, simple instrumentation |
| Impedimetry (EIS) | Impedance | Electrical resistance/ capacitance of interface | Label-free, real-time monitoring |
| Voltammetry | Current vs. Potential | Redox behavior during potential sweep | High sensitivity and selectivity (e.g., DPV) |
| Conductometry | Conductance | Ionic strength change in bulk solution | Simple, direct measurement |
The interplay between the bioreceptor and transducer, often enhanced with nanomaterials, defines the biosensor's mechanism. The following diagram details the operational principles of two common biosensor types: catalytic (e.g., enzyme-based) and affinity-based (e.g., antibody or aptamer-based).
Experimental Protocols for Biosensor Development
Protocol: Fabrication of a Nanomaterial-Modified Working Electrode
The sensitivity of modern biosensors is heavily dependent on the electrode's surface area and electronic properties. Nanomaterials are often used to modify the working electrode to enhance its performance [5] [3].
1. Aim: To fabricate a screen-printed carbon electrode (SPCE) modified with multi-walled carbon nanotubes (MWCNTs) and gold nanoparticles (AuNPs) to create a high-sensitivity platform for pathogen detection [7]. 2. Materials: * Screen-printed carbon electrode (SPCE) * Carboxyl-functionalized multi-walled carbon nanotubes (MWCNT-COOH) * Chloroauric acid (HAuCl₄) * Phosphate buffer saline (PBS, 0.1 M, pH 7.4) * N,N-Dimethylformamide (DMF) * Eppendorf tubes and micropipettes 3. Procedure: * MWCNT Dispersion: Disperse 1 mg of MWCNT-COOH in 1 mL of DMF and sonicate for 30 minutes to obtain a homogeneous black suspension. * Electrode Modification: Drop-cast 5 µL of the MWCNT suspension onto the working electrode area of the SPCE and allow it to dry at room temperature. * AuNP Electrodeposition: Immerse the MWCNT/SPCE in a 0.1 M PBS solution containing 1 mM HAuCl₄. Perform cyclic voltammetry between -0.2 V and +1.0 V (vs. Ag/AgCl reference) for 10 cycles at a scan rate of 50 mV/s to electrodeposit AuNPs. * Rinsing and Storage: Rinse the modified electrode (MWCNT-AuNP/SPCE) thoroughly with deionized water and store dry at 4°C when not in use.
Protocol: Immobilization of an Aptamer Bioreceptor
Aptamers are a popular choice for bioreceptors due to their stability and specificity. This protocol describes their covalent immobilization on a nanomaterial-modified electrode [1] [3].
1. Aim: To covalently immobilize a thiol-modified DNA aptamer onto a gold nanoparticle-modified electrode surface for the detection of a specific pathogen. 2. Materials: * MWCNT-AuNP/SPCE (from Protocol 3.1) * Thiol-modified DNA aptamer (e.g., specific for E. coli O157:H7) * N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) and N-Hydroxysuccinimide (NHS) * 6-Mercapto-1-hexanol (MCH) * Tris-EDTA (TE) buffer 3. Procedure: * Aptamer Preparation: Dilute the thiol-modified aptamer to 1 µM in TE buffer and reduce the thiol groups by incubating with Tris(2-carboxyethyl)phosphine (TCEP) for 1 hour. * Covalent Immobilization: Incubate the MWCNT-AuNP/SPCE with a mixture of 0.4 M EDC and 0.1 M NHS in water for 30 minutes to activate carboxyl groups on the MWCNTs. Then, drop-cast 10 µL of the reduced aptamer solution onto the activated surface and incubate in a humid chamber for 2 hours at 37°C. * Surface Blocking: To minimize non-specific binding, incubate the electrode with 1 mM 6-Mercapto-1-hexanol (MCH) for 30 minutes. This step passivates unoccupied gold sites. * Rinsing and Storage: Rinse the aptamer-functionalized biosensor with PBS to remove unbound aptamers. The biosensor can be stored in PBS at 4°C until use.
Protocol: Electrochemical Detection of Pathogen via Differential Pulse Voltammetry
This protocol outlines the use of the fabricated biosensor for the quantitative detection of a target pathogen using a sandwich assay format and DPV measurement [9].
1. Aim: To detect E. coli O157:H7 using an aptamer-based biosensor and a ferrocene-labeled reporter in a sandwich assay. 2. Materials: * Aptamer-functionalized biosensor (from Protocol 3.2) * Samples containing E. coli O157:H7 (concentration range 10² - 10⁸ CFU/mL) * Ferrocene-conjugated secondary aptamer or antibody * Electrochemical analyzer with DPV capability 3. Procedure: * Sample Incubation: Incubate the biosensor with 50 µL of the sample solution (or standard) for 20 minutes at room temperature to allow pathogen binding. * Sandwich Complex Formation: Rinse the sensor gently with PBS. Then, incubate it with 50 µL of the ferrocene-conjugated secondary detection probe for 15 minutes. * Electrochemical Measurement: After a final rinse, perform DPV measurement in a clean electrochemical cell containing 0.1 M PBS. The typical parameters are: potential window from 0 V to +0.5 V, pulse amplitude of 50 mV, and pulse width of 50 ms. * Data Analysis: The oxidation current peak of ferrocene (typically around +0.3 V) is measured. Plot the peak current against the logarithm of the pathogen concentration to generate a calibration curve for quantitative analysis.
The Scientist's Toolkit: Essential Research Reagents & Materials
The development and deployment of electrochemical biosensors for agricultural research rely on a specific set of reagents and materials. The following table details key components for building and experimenting with these devices.
Table 3: Essential Research Reagents and Materials for Biosensor Development
| Item | Function/Application |
|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, cost-effective sensor platforms with integrated working, reference, and counter electrodes. Ideal for field deployment [9]. |
| Carbon Nanotubes (CNTs) | Nanomaterials used to modify electrodes. They provide a high surface area, enhance electron transfer, and can be functionalized for bioreceptor immobilization [5] [3]. |
| Gold Nanoparticles (AuNPs) | Nanomaterials that improve conductivity and facilitate the stable immobilization of thiol-modified bioreceptors like antibodies or aptamers [7]. |
| Specific Bioreceptors | Engineered antibodies, aptamers, or DNA probes designed to bind with high affinity to a specific agricultural analyte (e.g., a fungal protein or mycotoxin) [1] [3]. |
| EDC & NHS Crosslinkers | Chemicals used for covalent immobilization of bioreceptors (especially those with carboxyl or amine groups) onto electrode surfaces [3]. |
| Electrochemical Redox Probes | Molecules such as Ferrocene or Hexaammineruthenium(III) chloride that act as labels to generate an amplified electrochemical signal in sandwich-type assays [9]. |
| Phosphate Buffered Saline (PBS) | A standard buffer used to maintain a stable pH and ionic strength during bioreceptor immobilization and electrochemical measurements [7]. |
Electrochemical biosensors have emerged as transformative analytical tools for precision agriculture, enabling the rapid, sensitive, and in-situ detection of vital plant nutrients, soil contaminants, and pathogen-derived biomarkers [10]. These sensors function at the interface of biology, chemistry, and material science, translating complex biological interactions into quantifiable electrical signals [11]. For agricultural researchers, the capability to monitor parameters such as soil contaminant levels or plant nitrogen status in real-time directly in the field represents a critical advancement over traditional laboratory-based methods, which are often hampered by complex pretreatment protocols and an inability to capture dynamic fluctuations [12] [13]. This technical guide provides an in-depth examination of three foundational electrochemical detection techniques—amperometry, voltammetry, and impedance spectroscopy—detailing their principles, implementations, and specific applications within agricultural research to support the development of next-generation smart farming systems.
Core Technique Principles and Agricultural Applications
Amperometry
2.1.1 Principle and Mechanism Amperometric sensors operate by applying a constant potential to an electrochemical cell and measuring the resulting Faradaic current generated from the oxidation or reduction of electroactive species at the electrode surface [13] [11]. The measured current is directly proportional to the concentration of the target analyte. Key performance characteristics include sub-micromolar detection limits and sub-second response times, making this technique uniquely suited for capturing transient chemical events in biological systems [13]. A significant advantage of modern amperometric systems is their excellent RC time constant properties (less than 100 μs), which provides a high signal-to-noise ratio essential for monitoring rapid enzymatic reactions or nutrient uptake kinetics in plants [13].
2.1.2 Agricultural Implementation and Protocols In agricultural settings, amperometric sensors are particularly valuable for monitoring inorganic nitrogen species crucial for plant health, such as nitrate (NO₃⁻) and nitric oxide (NO) [13]. The experimental protocol typically involves polarizing a micro- or nano-scale working electrode at a predetermined potential specific to the target analyte. For in-situ plant monitoring, needle-type amperometric sensors can be inserted directly into plant stems or root zones to sample xylem and phloem sap, enabling real-time tracking of dynamic nitrogen fluxes during nutrient uptake and stress responses [13].
A standard methodology involves the following steps:
- Electrode Preparation: Modify working electrodes with nanomaterials (e.g., graphene, carbon nanotubes) or catalytic layers to enhance sensitivity and selectivity toward the target nitrogen species [13] [11].
- Potential Optimization: Determine the optimal working potential for the target analyte using preliminary cyclic voltammetry scans to identify the oxidation/reduction peak potentials.
- Calibration: Perform standard additions of the analyte to establish a linear relationship between current response and concentration.
- In-Situ Measurement: Deploy the calibrated sensor in the agricultural environment (e.g., inserted into plant tissue, immersed in soil leachate) while applying the constant potential and recording the steady-state current.
Voltammetry
2.2.1 Principle and Mechanism Voltammetry encompasses a group of techniques that measure current while systematically varying the applied potential between working and reference electrodes [13]. Different voltammetric methods offer unique capabilities for agricultural sensing:
- Cyclic Voltammetry (CV): applies a linear potential ramp that reverses direction at a set switching potential, providing information about redox potentials and reaction mechanisms of electroactive species [13].
- Differential Pulse Voltammetry (DPV): applies small amplitude potential pulses superimposed on a linear ramp, enhancing sensitivity through background current rejection, making it ideal for detecting low-concentration analytes in complex matrices like soil samples [13].
- Linear Sweep Voltammetry (LSV): employs a single linear potential sweep, useful for identifying redox signatures of nitrogenous analytes in plant systems [13].
2.2.2 Agricultural Implementation and Protocols Voltammetric techniques find extensive application in detecting pesticides, pharmaceutical contaminants, and heavy metals in soil and agricultural products [12] [14]. The redox-active characteristics of these contaminants make them particularly amenable to voltammetric analysis. For example, DPV has been successfully employed for the simultaneous determination of multiple neutral nitrogen compounds in complex environmental samples [13].
A representative experimental workflow for detecting soil contaminants involves:
- Sample Collection: Obtain soil cores or leachates from agricultural fields with minimal pretreatment to preserve native conditions.
- Electrode Modification: Fabricate nanostructured electrodes (e.g., with metal-organic frameworks or molecularly imprinted polymers) to enhance specificity toward target contaminants [12] [15].
- Electrochemical Analysis:
- For CV: Sweep potential between predetermined limits (e.g., -1.0 V to +1.0 V) at scan rates typically between 10-100 mV/s.
- For DPV: Apply pulses with amplitudes of 25-50 mV and durations of 50-100 ms.
- Data Interpretation: Identify target contaminants based on their characteristic peak potentials and quantify concentrations through calibration curves.
Electrochemical Impedance Spectroscopy (EIS)
2.3.1 Principle and Mechanism Impedimetric biosensors detect subtle changes in the electrical properties (resistance and capacitance) at the electrode-electrolyte interface upon binding of target analytes [11]. EIS measurements involve applying a small amplitude sinusoidal AC potential across a frequency range and measuring the resulting current response to determine impedance. These systems are broadly classified into two categories:
- Faradaic EIS: Utilizes redox mediators (e.g., ferro/ferricyanide) and monitors changes in charge transfer resistance (Rct) upon target binding [11].
- Non-Faradaic EIS: Operates without redox couples, relying instead on changes in double-layer capacitance (Cdl) and interfacial dielectric properties, making it particularly advantageous for detecting analytes in their native state without sample alteration [11].
2.3.2 Agricultural Implementation and Protocols EIS has demonstrated exceptional utility for the label-free detection of plant pathogens, toxins, and disease biomarkers in oilseed crops [15]. Its ability to monitor binding events without requiring redox labels or extensive sample preparation makes it ideal for field-deployable agricultural diagnostics. For example, EIS-based sensors have been developed for early detection of fungal pathogens like Sclerotinia sclerotiorum in oilseed rape, achieving ultra-low detection limits through appropriate electrode functionalization [15].
A standard EIS protocol for plant pathogen detection includes:
- Biorecognition Immobilization: Functionalize gold or carbon-based working electrodes with specific capture elements (aptamers, antibodies) using thiol-gold chemistry or carbodiimide crosslinking [15] [11].
- Impedance Measurement: Apply a small AC voltage (5-10 mV) across a frequency range (typically 0.1 Hz to 100 kHz) at the open circuit potential.
- Data Modeling: Fit obtained spectra to equivalent electrical circuits (e.g., Randles circuit) to extract specific parameters (Rct, Cdl) that correlate with target concentration.
- Quantification: Monitor the increase in Rct or change in Cdl resulting from the binding of target pathogens to the electrode surface.
Table 1: Performance Comparison of Electrochemical Techniques in Agricultural Applications
| Technique | Detection Limit | Response Time | Key Agricultural Applications | Advantages |
|---|---|---|---|---|
| Amperometry | Sub-μM [13] | Sub-second [13] | Plant nitrogen species (NO₃⁻, NH₄⁺) monitoring | High temporal resolution, excellent for kinetic studies |
| Voltammetry | Variable (technique-dependent) | Seconds to minutes [13] | Pesticide detection, heavy metal screening in soils [12] | Identifies redox signatures, multi-analyte capability |
| Impedimetry | Femtomolar for pathogens [15] | Minutes [11] | Plant pathogen detection, soil contaminant monitoring [15] | Label-free detection, minimal sample preparation |
Experimental Design and Workflow
The successful implementation of electrochemical detection in agricultural research requires careful experimental design spanning from sensor fabrication to data interpretation. The following workflow diagram illustrates the integrated approach for real-time plant monitoring:
Figure 1: Integrated Workflow for Agricultural Electrochemical Sensing
Sensor Fabrication and Modification Strategies
Advanced sensor fabrication increasingly incorporates nanomaterials to enhance analytical performance. Nanostructured electrodes functionalized with graphene, carbon nanotubes, metal-organic frameworks, or metal nanoparticles provide increased surface area, improved electron transfer kinetics, and additional sites for immobilizing biorecognition elements [12] [11]. For agricultural applications requiring specificity toward biological targets, electrodes are modified with:
- Aptamers: Single-stranded DNA or RNA molecules that bind specific targets (pathogens, metabolites) with high affinity [15] [11].
- Antibodies: Immunorecognition elements for detecting plant pathogens or specific protein biomarkers [15].
- Molecularly Imprinted Polymers (MIPs): Synthetic polymers with tailor-made recognition sites for specific molecules, offering enhanced stability in harsh environmental conditions [11].
- Enzymes: Biological catalysts that provide specificity through substrate conversion to electroactive products [13].
Table 2: Research Reagent Solutions for Agricultural Electrochemical Sensing
| Reagent Category | Specific Examples | Function in Experimental Protocol |
|---|---|---|
| Electrode Materials | Gold microelectrodes, Screen-printed carbon, Platinum wire [11] | Signal transduction, providing conductive sensing platform |
| Nanomaterials | Graphene oxide, Carbon nanotubes, MXenes, Metal nanoparticles [12] [11] | Signal amplification, increased surface area, enhanced electron transfer |
| Biorecognition Elements | DNA aptamers, Antibodies, Enzymes (oxidoreductases) [15] [11] | Target specificity through molecular recognition |
| Redox Mediators | Ferro/ferricyanide, Methylene blue [11] | Facilitating electron transfer in faradaic systems |
| Stabilizing Matrices | Nafion, Chitosan, Self-assembled monolayers [15] | Biocompatible environments for biomolecule immobilization |
Signal Transduction Pathways in Electrochemical Biosensing
The fundamental signaling mechanisms in electrochemical biosensors involve a cascade of events from molecular recognition to measurable electrical outputs. The following diagram illustrates these transduction pathways:
Figure 2: Electrochemical Signal Transduction Pathways
Advanced Applications in Agricultural Research
Real-Time Plant Nutrient Monitoring
Electrochemical sensors have revolutionized precision nitrogen management by enabling real-time, in-situ detection of dynamically fluctuating nitrogen species in plants [13]. Breakthroughs in detection methodologies for inorganic nitrogen species (NO₃⁻, NH₄⁺, NO) have addressed critical gaps in traditional approaches limited by inadequate sensitivity and temporal resolution. Deployable miniaturized sensors now facilitate precision nitrogen management through direct integration with plant tissues or growth media, providing continuous data on nitrogen uptake kinetics and metabolic flux [13]. This capability is particularly valuable for optimizing fertilizer application schedules based on actual plant needs rather than predetermined regimens, significantly enhancing nitrogen use efficiency while reducing environmental pollution from agricultural runoff.
Case studies in maize and algal cultivation systems have demonstrated that electrochemical sensing technologies can support sustainable agricultural development by reducing excessive fertilizer use while maintaining or improving crop resilience and yield [13]. The integration of these sensor networks with Artificial Intelligence (AI) and Internet of Things (IoT) frameworks enables autonomous fertilization strategies tailored to real-time plant nitrogen demands, representing a paradigm shift in crop nutrient management [13].
Soil Contaminant Detection
Electrochemical sensors provide promising tools for rapid, sensitive, and selective detection of emerging contaminants (ECs) in soil, including pesticides, pharmaceuticals, heavy metals, and endocrine-disrupting compounds [12]. These contaminants pose significant threats to environmental and public health due to their diverse sources and complex environmental behaviors. Recent advances in electrochemical sensing have yielded enhanced detection limits, broader analyte ranges, and improved sensor stability under varying soil conditions [12].
Novel electrode materials and sensor designs have demonstrated particular effectiveness for monitoring soil pollution, with nanostructure-enhanced sensors showing remarkable improvements in specificity, sensitivity, and application potential [12]. The development of field-deployable electrochemical sensors for soil contaminant detection represents a critical advancement in environmental monitoring, enabling rapid assessment of soil health and prompt intervention when contamination is detected.
Plant Pathogen and Disease Diagnosis
Electrochemical biosensors have emerged as powerful tools for the early detection of diseases in economically important crops such as oilseed rape, soybean, and peanut [15]. Timely diagnosis is critical in agricultural management, as many pathogens exhibit latent infection phases or produce invisible metabolic toxins, leading to substantial yield losses before visible symptoms occur [15]. Innovations in nanomaterial-assisted electrochemical sensing have enabled the detection of pathogen DNA, enzymes, and toxins at ultra-low concentrations, providing a critical window for intervention before disease becomes established.
Specific applications include:
- Detection of Sclerotinia sclerotiorum (stem rot) in oilseed rape through identification of pathogen-specific biomarkers or secreted enzymes [15].
- Monitoring of Phakopsora pachyrhizi (soybean rust) infection by detecting effector proteins or changes in leaf physiology during the latent phase [15].
- Identification of aflatoxin-producing fungi in peanuts through toxin detection before visible symptoms manifest [15].
These applications demonstrate the transformative potential of electrochemical sensing for preventing crop losses and maintaining food security through early disease detection and targeted intervention.
Future Perspectives and Research Directions
The future development of electrochemical detection techniques for agricultural applications will likely focus on several key areas. Integration with artificial intelligence and machine learning algorithms will enhance data interpretation capabilities, enabling predictive analytics for plant health status and automated decision-making for agricultural management [13] [11]. Advances in wearable and minimally invasive sensor designs will facilitate long-term monitoring of plant physiological parameters without impairing growth or development [13]. The convergence of electrochemical sensing with wireless communication technologies and IoT networks will support the development of comprehensive agricultural monitoring systems capable of real-time, field-deployable disease surveillance and nutrient management [15].
Research efforts will also address current challenges in sensor stability, selectivity in complex matrices, and device miniaturization [15]. The exploration of biodegradable sensor materials represents an important direction for reducing environmental impact and ensuring sustainability in agricultural monitoring [15]. As these technologies mature, electrochemical detection techniques are poised to become cornerstone methodologies in smart agriculture, addressing global challenges in food security, environmental sustainability, and resource use efficiency.
In the rapidly evolving field of agricultural biotechnology, electrochemical biosensors have emerged as powerful analytical tools for monitoring pathogens, contaminants, and vital biomarkers across the food production chain. For researchers and drug development professionals implementing these technologies, a rigorous understanding of three fundamental performance metrics—sensitivity, selectivity, and limit of detection (LOD)—is paramount. These parameters collectively determine the reliability, accuracy, and practical utility of biosensing platforms in real-world agricultural applications, from precision farming to food safety monitoring [16] [17].
This technical guide provides an in-depth examination of these core metrics, establishing their theoretical foundations, practical measurement methodologies, and significance in agricultural research contexts. The content is structured to serve as both an educational resource for scientists new to biosensor development and a reference for experienced researchers validating analytical performance against regulatory standards.
Theoretical Foundations and Definitions
Sensitivity
Sensitivity quantifies the magnitude of signal change per unit change in analyte concentration. It represents the slope of the calibration curve, indicating how effectively a biosensor responds to minimal concentration variations of the target analyte [18]. In agricultural applications, high sensitivity is crucial for detecting trace-level contaminants like pesticides, mycotoxins, or bacterial pathogens in complex matrices such as soil, plant tissues, or food products [16].
Mathematically, sensitivity is defined as: [ S = \frac{\Delta S}{\Delta C} ] Where (S) is sensitivity, (\Delta S) is the change in sensor signal, and (\Delta C) is the change in analyte concentration.
Selectivity
Selectivity describes a biosensor's ability to distinguish the target analyte from interfering substances in a sample matrix. This characteristic is primarily determined by the specificity of the biological recognition element (enzyme, antibody, aptamer, or nucleic acid) toward its target [18]. In agricultural contexts with complex sample compositions, high selectivity ensures accurate measurements without false positives from chemically similar compounds or environmental interferents [19].
The related term specificity refers more narrowly to the capacity to identify an exact analyte in a mixture, while selectivity encompasses the broader ability to differentiate between multiple analytes [18].
Limit of Detection (LOD)
The Limit of Detection (LOD) is the lowest analyte concentration that can be reliably distinguished from a blank sample with a stated confidence level. It represents a fundamental parameter for assessing biosensor utility in early detection applications, such as identifying plant pathogens before visual symptoms appear [15] [20].
According to IUPAC definition, LOD is "the smallest solute concentration that a given analytical system can distinguish with reasonable reliability from a sample without analyte" [20]. The Limit of Quantification (LOQ), typically set at 10σ, represents the lowest concentration that can be quantitatively measured with acceptable precision and accuracy [18].
Table 1: Critical Performance Metrics for Electrochemical Biosensors
| Metric | Definition | Mathematical Expression | Agricultural Significance |
|---|---|---|---|
| Sensitivity | Change in signal per unit change in analyte concentration | ( S = \Delta S / \Delta C ) | Enables detection of low-level contaminants and pathogens |
| Selectivity | Ability to distinguish target from interfering substances | Not applicable | Ensures accuracy in complex matrices (soil, food, plant sap) |
| Limit of Detection (LOD) | Lowest detectable analyte concentration with statistical confidence | Typically ( 3\sigma_{blank}/S ) | Critical for early disease diagnosis and preventive intervention |
| Limit of Quantification (LOQ) | Lowest concentration measurable with acceptable precision | Typically ( 10\sigma_{blank}/S ) | Essential for quantitative monitoring of biomarkers |
Measurement Protocols and Methodologies
Determining Limit of Detection
The following procedural protocol outlines the standard method for determining LOD in label-free biosensors, adapted from international guidelines [20]:
Protocol: LOD Determination for Electrochemical Biosensors
Blank Measurement Preparation: Prepare a minimum of ( n_B ) replicates (typically ≥10) of blank solutions containing all components except the target analyte.
Signal Measurement: Measure the analytical response for each blank sample using the optimized biosensor platform.
Statistical Analysis: Calculate the mean (( \overline{yB} )) and standard deviation (( \sigma{blank} )) of the blank signals using Equations 1 and 2: [ \overline{yB} = \frac{\sum{j=1}^{nB} yj}{nB} ] [ \sigma{blank} = \sqrt{\frac{\sum{j=1}^{nB} (yj - \overline{yB})^2}{n_B - 1}} ]
Calibration Curve Construction: Prepare and analyze a minimum of 5 standard concentrations across the expected working range. Perform linear regression to establish the function ( y = aC + b ), where ( a ) is the sensitivity (slope) and ( b ) is the y-intercept.
LOD Calculation: Compute LOD using the formula: [ C{LOD} = \frac{3.3 \times \sigma{blank}}{a} ] The factor 3.3 corresponds to a 95% confidence level for both false positive and false negative rates [20].
The following diagram illustrates the statistical relationship between blank measurements, critical value, and LOD:
Assessing Sensitivity and Selectivity
Sensitivity Measurement Protocol:
- Generate a calibration curve with at least 5 concentration points across the analytical range
- Perform linear regression analysis to determine the slope (sensitivity) and correlation coefficient
- Report sensitivity with appropriate units (e.g., nA/μM, mV/decade) [18]
Selectivity Validation Protocol:
- Test biosensor response against structurally similar compounds and common matrix interferents
- Calculate selectivity coefficients for each potential interferent
- Evaluate performance in real samples with comparison to reference methods [19]
Performance Metrics in Agricultural Applications
Electrochemical biosensors in agricultural research must demonstrate robust performance across diverse and challenging environments. The table below summarizes reported performance metrics for various agricultural applications:
Table 2: Performance Metrics of Electrochemical Biosensors in Agricultural Applications
| Application | Target Analyte | Sensitivity | Selectivity Assessment | LOD | Reference Technique |
|---|---|---|---|---|---|
| Plant Pathogen Detection [15] | Sclerotinia sclerotiorum (Stem Rot) | Not specified | Demonstrated against other soil fungi | Early detection before vascular invasion | Visual inspection |
| Poultry Safety [21] | Salmonella spp., E. coli | Not specified | Differentiated from other enteric bacteria | Enables proactive flock management | Culture methods, PCR |
| GMO Screening [22] | EPSPS, PAT, Cry genes | Not specified | Specific DNA hybridization | Meets EU 0.9% threshold requirement | Multiplex qPCR |
| Viral Disease Monitoring [23] | Tobacco Mosaic Virus | Enhanced by AuNPs, MOFs | Minimal cross-reactivity | Significant improvement over ELISA | PCR, ELISA |
| Soil Nutrient Management [17] | Macronutrients (N, P, K) | Varies by ionophore | Ion-selective membranes | Sufficient for precision agriculture | Laboratory analysis |
Research Reagent Solutions for Biosensor Development
The following reagents and materials are essential for developing high-performance electrochemical biosensors for agricultural applications:
Table 3: Essential Research Reagents for Biosensor Development
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Gold Nanoparticles (AuNPs) | Enhance electron transfer, increase surface area for bioreceptor immobilization | Pathogen detection [24], viral disease monitoring [23] |
| Graphene Oxide (GO) | Provides large surface area with functional groups for stable probe immobilization | Contaminant monitoring, nutrient sensing [24] |
| Specific Antibodies | Immunological recognition elements for selective target binding | Pathogen detection [21], protein biomarker analysis |
| DNA/Aptamer Probes | Nucleic acid-based recognition with high specificity and stability | GMO detection [22], viral pathogen identification |
| Molecularly Imprinted Polymers (MIPs) | Synthetic recognition elements with high stability | Pesticide detection, small molecule analysis [24] |
| Ion-Selective Membranes | Enable selective detection of specific ions | Soil nutrient monitoring [17] |
Technological Integration and Emerging Approaches
The convergence of electrochemical biosensing with advanced materials and digital technologies represents the future of agricultural monitoring. The following diagram illustrates the integrated workflow from sensing to decision support:
Recent advances incorporate artificial intelligence and machine learning to enhance the interpretation of biosensor data, improving both the reliability of measurements and the predictive capabilities for agricultural management [16] [24]. The integration with Internet of Things (IoT) platforms enables real-time monitoring of crop health, soil conditions, and food contamination risks across distributed agricultural operations [16] [23].
Smartphone-integrated electrochemical devices represent a particularly promising development, combining laboratory-grade analysis with field-portable operation. These systems leverage the computational power, connectivity, and imaging capabilities of smartphones to create comprehensive mobile laboratories for on-site testing [24].
Sensitivity, selectivity, and limit of detection form the essential triad of performance metrics that dictate the practical utility of electrochemical biosensors in agricultural research and applications. As the field advances toward increasingly miniaturized, integrated, and intelligent monitoring systems, rigorous characterization and optimization of these parameters remains fundamental to transforming agricultural practices through precision management, early pathogen detection, and enhanced food safety assurance. The ongoing development of standardized protocols for evaluating these metrics will facilitate more meaningful comparisons between technologies and accelerate the translation of research innovations into practical agricultural solutions.
The Evolution from Laboratory Tools to Field-Deployable Agricultural Sensors
The transition of electrochemical biosensors from sophisticated laboratory instruments to robust field-deployable tools represents a paradigm shift in agricultural monitoring. This evolution is characterized by fundamental advances in nanomaterials engineering, bioreceptor stability, device miniaturization, and system integration, enabling direct detection of pathogens, toxins, and stress biomarkers in complex agricultural matrices. This whitepaper examines the technical trajectory of these sensing platforms, highlighting critical innovations in interface design, signal transduction, and data interoperability that support their integration within smart agricultural systems. The analysis further details standardized experimental protocols for performance validation and projects future development trajectories focused on artificial intelligence-driven interpretation and sustainable sensor architectures.
Fundamental Principles and Core Components
Electrochemical biosensors are analytical devices that integrate a biological recognition element with an electrochemical transducer to convert a biological event into a quantifiable electrical signal [5] [2]. The core architecture comprises bioreceptors (e.g., enzymes, antibodies, aptamers, nucleic acids) that provide selective binding to the target analyte, a transducer surface (typically an electrode) where the biochemical interaction occurs, and the electronic system that processes the signal into a readable output [5]. The significant advantage of electrochemical detection lies in its direct conversion of biological interaction to an electronic signal, enabling high sensitivity, minimal power requirements, and inherent compatibility with miniaturized, portable form factors [5] [25].
Table 1: Core Components of an Electrochemical Biosensor
| Component | Function | Common Materials & Examples |
|---|---|---|
| Bioreceptor | Provides selective binding to the target analyte | Enzymes (Glucose Oxidase), Antibodies, DNA/Aptamers, Whole Cells [16] [25] |
| Transducer | Converts the biological event into a measurable electrical signal | Gold, Carbon, or Platinum Electrodes; often nanomaterial-modified (e.g., Graphene Oxide, Gold Nanoparticles) [5] [25] |
| Electronics | Amplifies, processes, and displays the electrical signal | Potentiostats, custom ICs, integrated with microcontrollers and wireless communication modules [5] [25] |
The performance of these sensors is critically dependent on the precise control of the interface architecture at the nanoscale, where the interplay between surface functionalization, the chosen transducer principle, and the suppression of non-specific interactions determines ultimate sensitivity and specificity [5].
Figure 1: Core signaling pathway of an electrochemical biosensor, illustrating the conversion of a biological binding event into actionable data via a transducer.
Performance Metrics and Experimental Protocols
Validating sensor performance requires standardized methodologies to assess key metrics critical for both laboratory research and field application. The following protocols and metrics are essential for benchmarking.
Key Performance Metrics (KPIs)
Table 2: Key Performance Metrics for Agricultural Electrochemical Biosensors
| Metric | Definition | Target for Field Deployment |
|---|---|---|
| Limit of Detection (LOD) | The lowest analyte concentration that can be reliably distinguished from a blank | Ultra-low concentrations (e.g., pathogen DNA at femtomolar levels, toxins at μg/kg) [3] |
| Linear Range | The concentration interval over which the sensor response is linearly proportional to analyte concentration | Covers the clinically/agronomically relevant concentration span [3] |
| Selectivity/Specificity | The ability to detect the target analyte without interference from similar substances or matrix components | High specificity in complex matrices like soil extracts, plant sap, or food samples [16] |
| Stability | The ability to maintain performance over time and under storage conditions | Long-term stability (weeks to months) under variable temperature/humidity [16] |
| Reproducibility | The precision of measurements across different sensors or batches | Low coefficient of variation (<5-10%) between manufactured units [5] |
Detailed Experimental Protocol for Sensor Validation
The following protocol outlines a standard procedure for characterizing an electrochemical aptasensor for pathogen detection, adaptable for other targets.
Aim: To characterize the performance of a nanomaterial-modified electrochemical biosensor for the detection of Sclerotinia sclerotiorum DNA in a spiked plant extract sample.
Materials & Reagents:
- Working Electrode: Screen-printed carbon electrode (SPCE) modified with graphene oxide (GO) and gold nanoparticles (AuNPs) [25].
- Bioreceptor: DNA aptamer specific to S. sclerotiorum, thiol-modified for immobilization.
- Target Analyte: Synthetic oligonucleotide of the target pathogen DNA sequence.
- Electrochemical Probe: Potassium ferricyanide/ferrocyanide ([Fe(CN)₆]³⁻/⁴⁻) in buffer.
- Matrix: Phosphate Buffered Saline (PBS, pH 7.4) for initial tests, followed by a filtered extract from healthy oilseed rape leaves to simulate a real sample matrix [3].
Methodology:
- Electrode Modification:
- Nanomaterial Deposition: Drop-cast 5-10 μL of a GO dispersion onto the SPCE surface and dry under nitrogen. Electrochemically reduce GO to conductive reduced graphene oxide (rGO) by performing cyclic voltammetry (CV) in a suitable buffer.
- AuNP Electrodeposition: Immerse the rGO/SPCE in a solution of HAuCl₄ and apply a constant potential to electrodeposit AuNPs, enhancing surface area and facilitating thiol binding [25].
- Aptamer Immobilization: Incubate the modified electrode with a 1 μM solution of the thiolated aptamer for 12-16 hours. Passivate any remaining gold surface with 6-mercapto-1-hexanol (MCH) to minimize non-specific binding.
Electrochemical Measurement:
- Perform electrochemical impedance spectroscopy (EIS) in the presence of the [Fe(CN)₆]³⁻/⁴⁻ redox probe after each modification step (bare SPCE, rGO/SPCE, AuNP/rGO/SPCE, aptamer/MCH/AuNP/rGO/SPCE) to monitor the increase in electron transfer resistance (Rₑₜ), confirming successful layer-by-layer assembly.
- For quantitative detection, incubate the functionalized sensor with standard solutions of the target DNA (e.g., 1 pM to 100 nM) or spiked plant extracts for a fixed time (e.g., 20-30 minutes).
- Record the EIS or differential pulse voltammetry (DPV) signal after each incubation. The binding of the target DNA increases Rₑₜ or causes a change in current, which is proportional to the target concentration.
Data Analysis:
- Plot the change in Rₑₜ or current against the logarithm of the target concentration.
- Perform a linear regression analysis on the linear portion of the curve to establish the calibration plot.
- Calculate the LOD as 3.3 × (standard deviation of the blank/slope of the calibration curve).
The Trajectory from Laboratory to Field
The evolution of these sensors from lab to field is driven by specific technological breakthroughs that address the challenges of complexity, stability, and usability.
Miniaturization and Integration
Early laboratory biosensors relied on bulky, three-electrode systems connected to benchtop potentiostats. The adoption of screen-printed electrodes (SPEs), which integrate working, reference, and counter electrodes on a single, disposable chip, was a pivotal step toward portability and low-cost mass production [25]. Further integration with microfluidics (Lab-on-a-Chip, LoC) automates sample handling and reduces reagent volumes, making the device suitable for raw, minimally processed agricultural samples [25].
Nanomaterials for Enhanced Sensitivity
The incorporation of nanomaterials is a cornerstone of this evolution. Gold nanoparticles (AuNPs) and graphene oxide (GO) are engineered into electrode surfaces to provide a high surface-to-volume ratio for increased bioreceptor loading, enhanced electrical conductivity for faster electron transfer, and catalytic properties for signal amplification. This nanomaterial-driven enhancement allows for detection at ultra-low concentrations, which is crucial for identifying latent infections before symptoms appear [3] [25].
Connectivity and Data Interoperability
Modern field-deployable sensors transcend mere detection. Integration with smartphone-based potentiostats provides computational power, intuitive user interfaces, and cloud connectivity, transforming the sensor into a node in a larger Internet of Things (IoT) network [25]. This enables real-time data transmission to agricultural decision-support systems, facilitating immediate interventions and contributing to large-scale, data-driven pest and disease models [3] [16].
Figure 2: Workflow evolution from laboratory-based analysis to connected field-deployment.
Implementation Challenges and Material Solutions
Despite significant progress, the full deployment of electrochemical biosensors in agriculture faces several hurdles. The research community is actively developing innovative material and strategic solutions to address these challenges.
Table 3: Key Implementation Challenges and Emerging Solutions
| Challenge | Impact on Deployment | Emerging Solutions |
|---|---|---|
| Matrix Interference | Complex agricultural samples (soil, sap) cause fouling and false signals, reducing accuracy. | - Use of robust antifouling membranes (e.g., hydrogels) [16].- Advanced surface chemistries to repel non-specific adsorption [5].- Integration of microfluidics for sample filtration/separation [25]. |
| Device Stability & Calibration | Sensitivity drifts over time due to bioreceptor denaturation, requiring frequent re-calibration. | - Development of synthetic bioreceptors (MIPs, engineered aptamers) with higher stability [25].- Exploration of reagent-free sensing mechanisms.- On-device calibration algorithms. |
| Standardization & Scalability | Lack of uniform manufacturing and testing protocols hinders regulatory approval and mass production. | - Adoption of scalable fabrication techniques like screen printing and inkjet printing [25].- Development of consensus performance standards and validation protocols for agri-food targets [16]. |
The Scientist's Toolkit: Research Reagent Solutions
The development and implementation of advanced electrochemical biosensors rely on a suite of specialized reagents and materials.
Table 4: Essential Research Reagent Solutions for Agricultural Biosensing
| Research Reagent | Function in Experimental Protocol |
|---|---|
| Screen-Printed Electrodes (SPEs) | Provide a disposable, miniaturized, and reproducible platform for sensor fabrication, forming the core of portable devices [25]. |
| Gold Nanoparticles (AuNPs) & Graphene Oxide (GO) | Nanomaterials used to modify electrode surfaces, enhancing sensitivity and signal-to-noise ratio by increasing surface area and facilitating electron transfer [25]. |
| Thiol-modified Aptamers | Serve as stable, synthetic bioreceptors. The thiol group allows for covalent, oriented immobilization on gold surfaces (e.g., AuNPs), improving binding efficiency and sensor consistency [3] [25]. |
| Redox Probes (e.g., [Fe(CN)₆]³⁻/⁴⁻) | Used in electrochemical techniques like EIS and CV to monitor the success of electrode modification and to transduce the biorecognition event into a measurable electrical signal [5]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymer scaffolds with tailor-made cavities for a specific analyte. They act as artificial antibodies, offering superior stability and lower cost than biological receptors for targets like pesticide residues [25]. |
The evolution of agricultural biosensors is continuing along several innovative trajectories. Artificial Intelligence (AI) and Machine Learning (ML) are being integrated to handle complex data interpretation, compensating for sensor drift and environmental variability to improve prediction accuracy [3] [16]. A growing emphasis on sustainability is driving research into biodegradable sensor substrates and green manufacturing processes to minimize environmental impact [3]. Finally, the future lies in closed-loop systems, where sensor data automatically triggers agricultural actuators, such as initiating precision spraying or irrigation in real-time, fully realizing the promise of data-driven, sustainable precision agriculture [16].
In conclusion, the journey of electrochemical biosensors from laboratory tools to field-deployable assets is a testament to interdisciplinary innovation. Through advances in nanotechnology, materials science, and microelectronics, these sensors are poised to become ubiquitous tools for safeguarding crop health, optimizing resource use, and strengthening global food security.
From Lab to Field: Sensor Applications for Crop and Soil Monitoring in Precision Agriculture
Oilseed crops are vital components of global agriculture, supplying over 80% of edible oils and 40% of biofuel feedstock worldwide [3] [15]. However, their productivity is consistently threatened by devastating pathogens and the carcinogenic aflatoxins they can produce, leading to substantial economic losses and serious food safety concerns. Timely diagnosis is critical in disease management, as many pathogens exhibit latent infection phases where they colonize plant tissues without visible symptoms [3]. This technical guide explores the application of electrochemical biosensors as promising tools for early detection of oilseed pathogens and aflatoxins, focusing on their operational mechanisms, performance metrics, and implementation protocols within precision agriculture frameworks.
Major Oilseed Pathogens and Aflatoxins: Economic and Health Impacts
The following table summarizes the key pathogens affecting major oilseed crops, their detection windows, and the associated economic and health impacts.
Table 1: Major Oilseed Pathogens and Aflatoxins: Characteristics and Impacts
| Pathogen/Toxin | Primary Host(s) | Key Characteristics | Economic/Health Impact | Optimal Detection Window |
|---|---|---|---|---|
| Sclerotinia sclerotiorum (Stem Rot) | Oilseed Rape, Canola | Fungus survives in soil for years as sclerotia; produces ascospores dispersed by wind [3]. | Annual global yield loss: 15-20%; economic losses >$5 billion. In Canada (2022), led to 18% production decline in Manitoba [3]. | Early appearance of water-soaked lesions on stems, before hyphal invasion of vascular tissues [3]. |
| Phakopsora pachyrhizi (Soybean Rust) | Soybean | Airborne urediniospores can travel >1000 km/month; degrades thylakoid membranes within 72h [3]. | 2023 Brazil epidemic caused loss of 2.1M tons ($1.4B); latent infections can colonize 40% of leaf area before symptoms [15]. | Latent infection phase, before visible symptoms manifest [15]. |
| Sclerotium rolfsii (White Mold) | Peanut | Melanized sclerotia withstand 45°C soil temps, remain viable for 5-8 years; secretes cell-wall degrading enzymes [3] [15]. | Causes 20-50% yield loss in wet ecosystems; pod weight loss of 35-50% [3]. | Not specified in search results. |
| Aflatoxin B1 (AFB1) | Peanut, Soybean, Maize | Potent carcinogen produced by Aspergillus flavus and A. parasiticus; stable during processing [26] [27]. | Contributed to $320M in export rejections from India's Telangana region (2024); classified as Class 1 carcinogen; synergistically increases liver cancer risk with Hepatitis B [3] [26]. | Pre-harvest and post-harvest stages; critical in stored grains and edible oils [26]. |
Electrochemical Biosensors: Core Technologies and Performance
Electrochemical biosensors integrate a biological recognition element with an electrode transducer, converting a target-analyte interaction into a quantifiable electrical signal [16]. The performance of these sensors is significantly enhanced by nanomaterials and various biorecognition elements.
Table 2: Core Components and Performance of Nanomaterial-Based Electrochemical Biosensors
| Sensor Component | Function & Role | Key Innovations & Examples | Reported Performance Gains |
|---|---|---|---|
| Nanostructured Electrodes | Enhance surface area, conductivity, and electron transfer rates; improve loading capacity for bioreceptors [3] [7]. | Use of multi-walled carbon nanotubes (MWCNTs), graphene, metal nanoparticles (e.g., gold), and metal-organic frameworks (MOFs) [7]. | Increased signal-to-noise ratio, lower limits of detection (LOD), and higher sensitivity in complex matrices [3] [7]. |
| Biorecognition Elements | Provide specificity by binding to the target analyte (pathogen DNA, toxin, enzyme) [3] [16]. | Aptamers: Single-stranded DNA/RNA oligonucleotides (e.g., structure-switching aptamer for AFB1) [28].Antibodies: Immunological recognition [3].Molecularly Imprinted Polymers (MIPs): Artificial antibody mimics [26]. | High specificity; aptamers offer advantages of stability and lower production cost compared to antibodies [26] [28]. |
| Signal Amplification | Augments the electrochemical response from the binding event, enabling ultra-low concentration detection [3] [7]. | Techniques include catalytic nanomaterials, enzymatic amplification, and cascading reactions [3]. | Enables detection of pathogen DNA, enzymes, and toxins at ultra-low concentrations (picomolar to femtomolar) [3]. |
Experimental Protocols for Biosensor Deployment
Protocol 1: Paper-based Ratiometric Aptasensor for Aflatoxin B1 Detection
This protocol outlines the procedure for constructing a disposable paper-based electrochemical biosensor for AFB1, leveraging a structure-switching aptamer for specificity and a ratiometric measurement for accuracy [28].
- Principle: The sensor uses a methylene blue (MB)-tagged DNA aptamer co-assembled with a ferrocene (Fc) internal reference on a paper-based electrode. In the presence of AFB1, the aptamer folds, distancing the MB tag from the electrode surface and reducing its current signal. The current ratio of MB to Fc provides a robust, internally-corrected measurement [28].
- Materials:
- Paper-based working electrode (e.g., screen-printed carbon electrode)
- AFB1-specific DNA aptamer tagged with Methylene Blue (MB)
- Internal reference probe tagged with Ferrocene (Fc)
- Differential Pulse Voltammetry (DPV) apparatus
- Buffer solutions for immobilization and washing
- Step-by-Step Procedure:
- Electrode Co-assembly: Immobilize the MB-tagged aptamer and the Fc-tagged reference probe onto the paper-based electrode surface.
- Sample Incubation: Apply the sample extract (e.g., from groundnuts or edible oil) to the sensor surface and incubate to allow AFB1-aptamer binding.
- Electrochemical Measurement: Perform DPV measurements in a suitable buffer.
- Signal Analysis: Record the oxidation peak currents for both MB (IMB) and Fc (IFc).
- Quantification: Calculate the ratiometric signal (IMB / IFc). The signal decreases proportionally with increasing AFB1 concentration. Compare to a calibration curve for quantification.
- Performance Metrics: This sensor demonstrated a linear detection range of 20.0 to 1000.0 ng mL⁻¹ and a limit of detection (LOD) of 7.6 ng mL⁻¹. It showed high specificity and stability, with results correlating well with standard UPLC−MS/MS analysis in real samples [28].
Protocol 2: Smartphone-based Digital Image Colorimetry with Nanobiosensor
This protocol describes a colorimetric method integrating a bio-nanoparticle sensor with smartphone technology for sensitive AFB1 detection, suitable for resource-limited settings [29].
- Principle: Curcumin-functionalized ZnO nanoparticles form the sensing platform. AFB1 binding induces a color change in the nanoparticle-curcumin complex. Dispersive liquid-liquid microextraction (DLLME) pre-concentrates AFB1 from the sample. A smartphone camera in a portable light-box captures the color, which is analyzed via a colorimetric app [29].
- Materials:
- Bio-synthesized ZnO Nanoparticles functionalized with curcumin
- DLLME solvents: Chloroform (extraction solvent) and acetonitrile (disperser solvent)
- Smartphone with colorimetry application
- Portable colorimetric box with standardized LED lighting (45° angle, 4 LEDs recommended)
- Step-by-Step Procedure:
- Sample Pre-concentration: Perform DLLME on the liquid food sample (e.g., oil extract) using chloroform and acetonitrile to isolate and concentrate AFB1.
- Sensor Incubation: Re-dissolve the extracted AFB1 and mix with the ZnO-NPs/curcumin nano-biosensor complex at the optimized ratio (2:1 curcumin to NPs) and pH (9.44).
- Reaction: Allow the reaction to proceed for 2-3 minutes.
- Image Capture: Place the solution in the portable box and capture an image using the smartphone camera under standardized lighting.
- Color Channel Analysis: Analyze the image using the green (G) color channel intensity or the G/R ratio, which shows the best linear response to AFB1 concentration.
- Performance Metrics: This method achieved a remarkably low LOD of 0.09 μg/kg and a linear range of 0–1 μg/L, with high recoveries (89.8–94.2%) in baby food samples [29].
Visualizing Biosensor Workflows and Pathways
The following diagram illustrates the logical workflow and key decision points in developing and deploying an electrochemical biosensor for agricultural pathogens.
The Scientist's Toolkit: Essential Research Reagents and Materials
This table catalogs key reagents and materials essential for developing and implementing electrochemical biosensors for oilseed pathogen detection.
Table 3: Key Research Reagent Solutions for Biosensor Development
| Item/Category | Specific Examples | Function in Biosensing |
|---|---|---|
| Biorecognition Elements | DNA/Aptamers (e.g., structure-switching aptamer for AFB1) [28]; Antibodies (vs. pathogens); Molecularly Imprinted Polymers (MIPs) [26] | Provides high specificity and selectivity for the target analyte (pathogen, toxin). |
| Nanomaterials for Electrodes | Multi-walled Carbon Nanotubes (MWCNTs) [7]; Gold Nanoparticles (AuNPs) [7]; Graphene; Metal-Organic Frameworks (MOFs) [7] | Enhances electrode conductivity, surface area, and signal amplification. Improves biosensor sensitivity and LOD. |
| Electrochemical Tags | Methylene Blue (MB) [28]; Ferrocene (Fc) [28] | Redox reporters that generate measurable current changes upon target binding in voltammetric sensors. |
| Sensor Substrates | Screen-Printed Electrodes (SPEs); Paper-based electrodes [28] | Provides a low-cost, disposable, and portable platform for single-use field-deployable sensors. |
| Signal Transduction Equipment | Potentiostat; Smartphone with colorimetry app [29] | Measures and interprets the electrochemical (current, impedance) or optical (color change) signal. |
Challenges and Future Research Directions
Despite significant advancements, the translation of laboratory biosensor prototypes to field-deployable tools faces several hurdles. A major challenge is the lack of real-world validation; a systematic review found that only 1 out of 77 studies tested biosensors on naturally contaminated food samples, with most relying on artificially spiked samples [7]. Other challenges include signal interference from complex plant matrices, limited device miniaturization, and the absence of standardized detection protocols [3] [16].
Future research should focus on:
- Validation and Standardization: Establishing protocols for testing with naturally contaminated samples and aligning performance metrics with international standards (ISO, FAO, FDA) [7].
- AI and IoT Integration: Leveraging artificial intelligence for data interpretation and integrating sensors into IoT networks for real-time, spatially-resolved disease surveillance in smart agriculture systems [3] [16].
- Material Innovation: Developing more stable bioreceptors and biodegradable sensor materials to enhance shelf-life and reduce environmental impact [3].
- Multiplexing: Creating sensors capable of simultaneously detecting multiple pathogens or toxins in a single assay to provide comprehensive crop health diagnostics [16] [4].
The transition to precision agriculture necessitates a shift from retrospective to real-time diagnostic tools for monitoring plant physiochemical signals. This whitepaper examines the principles and methodologies of sap analysis and nutrient solution monitoring, framing them within the advancing context of electrochemical biosensor technology. These tools provide a dynamic window into the plant's physiological status, enabling detection of active, soluble nutrients and signaling molecules with high temporal resolution. While sap analysis offers a snapshot of the mobile nutrients within the vascular system, electrochemical biosensors are emerging as a revolutionary technology for the in-situ and real-time detection of specific plant signaling molecules and stressors. This technical guide details standardized protocols, data interpretation frameworks, and the integration of these tools into a comprehensive sensor-based decision support system for research and development.
Plant health and productivity are governed by complex physiological processes influenced by environmental conditions and genetic makeup. Traditional plant analysis methods, such as tissue testing, provide a historical record of nutrient accumulation but lack the temporal resolution to capture dynamic changes in nutrient mobility and stress signaling. The limitations of tissue analysis have driven the development of advanced monitoring techniques that offer real-time or near-real-time insights [30].
Electrochemical biosensors represent a paradigm shift in this domain. These devices combine a biological recognition element with an electrochemical transducer, offering convenient methods for in-situ and real-time detection of plant signaling molecules due to their easy operation, high sensitivity, and high selectivity [31] [32]. This whitepaper explores how established sap analysis practices and cutting-edge electrochemical sensors collectively contribute to a deeper, more immediate understanding of plant physiochemistry, providing researchers with powerful tools for optimizing plant health and productivity.
Plant Sap Analysis: Principles and Methodologies
What is Plant Sap Analysis?
Plant sap analysis is a diagnostic technique that measures the concentration of soluble nutrients present in the vascular tissues (xylem and phloem) of a plant. Unlike traditional tissue analysis, which involves drying and grinding entire plant parts to measure total accumulated nutrients, sap analysis extracts the liquid component from fresh plant tissues to assess the nutrients that are actively circulating [33] [30]. This provides a near real-time assessment of nutrient availability within the plant, allowing for the detection of imbalances often weeks before visual symptoms manifest [33].
Comparative Analysis: Sap vs. Tissue Testing
The choice between sap analysis and traditional tissue testing depends on the specific research or monitoring objectives. The following table summarizes their key differences.
Table 1: Comparative Analysis of Sap Analysis and Standard Tissue Testing
| Aspect | Sap Analysis | Standard Tissue Analysis |
|---|---|---|
| Sample Type | Extracted sap from fresh plant tissues (e.g., leaves, petioles) | Dried and ground plant tissues (e.g., leaves, stems) |
| Nutrient Measurement | Measures nutrients in the plant's vascular system, reflecting current availability | Measures total accumulated nutrients, including those structurally bound in tissues |
| Turnaround Time | Rapid results, often within hours to a few days | Longer processing time, typically several days to a week |
| Detection Sensitivity | Can detect nutrient imbalances before visual symptoms appear | May not detect deficiencies until they manifest visibly |
| Data Interpretation | Requires expertise due to variability influenced by environmental factors | More standardized interpretation with established sufficiency ranges |
| Nutrient Mobility Insight | Provides information on nutrient mobility by comparing young and old leaves | Offers a cumulative view but less insight into real-time nutrient movement |
| Environmental Sensitivity | Results can be affected by time of day, plant hydration, and conditions | Less sensitive to immediate environmental fluctuations [30] |
Key Experimental Protocol for Sap Analysis
A reliable sap analysis protocol is critical for generating accurate and reproducible data. The following methodology outlines the key steps from sample collection to data interpretation.
1. Sample Collection:
- Selection: Identify the target crop and the specific plant part to be sampled (e.g., petioles, leaf midribs). Consistency in selection is paramount.
- Timing: Collect samples at the same time of day, typically in the early morning, to minimize diurnal fluctuations in nutrient concentrations.
- Handling: Use linear pressure-based sap extractors that avoid mastication, heat, acids, or solvents to maintain the integrity of the leaf and the sap's chemical profile [33]. Immediately place samples in sealed bags and store on ice.
2. Sample Preparation & Shipping:
- To preserve sample integrity, ship them overnight to the analytical laboratory using pre-arranged cool shipping programs [33].
- Laboratories typically extract sap through controlled linear pressure, applying different pressures for different crop types to ensure leaves and cellular structures remain intact [33].
3. Laboratory Analysis:
- The extracted sap is analyzed for a comprehensive suite of parameters. Standard analyses include:
- Macronutrients: Nitrogen (as Nitrate and Ammonia), Phosphorus (P), Potassium (K), Calcium (Ca), Magnesium (Mg), Sulfur (S)
- Micronutrients: Boron (B), Copper (Cu), Iron (Fe), Manganese (Mn), Molybdenum (Mo), Zinc (Zn)
- Other Elements: Sodium (Na), Chlorine (Cl), Aluminum (Al), Nickel (Ni)
- Physicochemical Indices: Brix (soluble sugar content), pH, and Electrical Conductivity (EC) [33]
4. Data Interpretation:
- New vs. Old Leaf Comparison: A powerful feature of sap analysis is comparing nutrient levels in new and old growth. For mobile nutrients (e.g., Nitrogen, Potassium), a higher concentration in new leaves indicates remobilization from older leaves, suggesting insufficient root uptake. Conversely, an excess is indicated by higher concentrations in old leaves [33].
- Contextualization: Results must be interpreted considering crop type, growth stage, and environmental conditions to make accurate nutrient management recommendations.
The workflow below summarizes the key steps involved in the sap analysis process.
Electrochemical Biosensors for Advanced Monitoring
Principles and Sensor Types
Electrochemical biosensors are ideal for bridging the gap between sap analysis and continuous, in-situ monitoring. These sensors integrate a biological recognition element (e.g., enzyme, antibody, DNA/aptamer, whole cell) with an electrochemical transducer that converts a biological interaction into a quantifiable electrical signal [16]. The development of in-situ and real-time detection capabilities for plant signaling molecules is considered a key breakthrough for botanical research and agricultural technology [31].
Plant signaling molecules detected by these sensors can be broadly categorized as:
- Plant Messenger Signaling Molecules: Calcium ions (Ca²⁺), hydrogen peroxide (H₂O₂), Nitric oxide (NO) [31] [32].
- Plant Hormone Signaling Molecules: Auxin (IAA), salicylic acid, abscisic acid, cytokinin, jasmonic acid, gibberellins, brassinosteroids, strigolactone, and ethylene [31] [32].
Research Reagent Solutions for Biosensor Development
The fabrication of high-performance electrochemical biosensors relies on a suite of specialized materials and reagents. The following table details essential components used in this field.
Table 2: Key Research Reagent Solutions for Electrochemical Biosensor Development
| Reagent / Material | Function / Application | Specific Examples |
|---|---|---|
| Nanomaterials | Enhance electrode surface area, electron transfer kinetics, and overall sensitivity/selectivity. | Various nanomaterials are applied to enhance electrode detection [31]. |
| Biorecognition Elements | Provide specificity by binding to the target analyte. | Enzymes [16], Antibodies (Ab) [16], DNA/Aptamers [16], Whole cells (e.g., engineered E. coli) [34]. |
| Immobilization Matrices | Entrap and stabilize biorecognition elements on the transducer surface. | Alginate-based hydrogels (for whole-cell immobilization) [34]. |
| Electrode Materials | Serve as the solid support and transducer. | Glassy Carbon Electrode (GCE), carbon fibers, stainless steel (SS) wire, Indium Tin Oxide (ITO), SS sheets [31]. |
| Electrochemical Mediators | Shuttle electrons between the biorecognition element and the electrode surface. | Non-enzymatic nanoceria tag [16]. |
Experimental Protocol for a Whole-Cell Biosensor
Whole-cell biosensors utilize living microorganisms, genetically engineered to produce a measurable signal in response to specific stimuli, such as volatile organic compounds (VOCs) emitted by stressed or infected plants [34]. The following is a detailed protocol for creating such a sensor for detecting crop spoilage.
1. Bacterial Strain Preparation:
- Obtain genetically engineered luminescent bioreporter bacterial strains (e.g., E. coli TV1061 or DnaK strain containing a plasmid with a promoter fused to the luxCDABE operon) [34].
- Streak bacterial stocks on Luria-Bertani (LB) agar plates supplemented with an appropriate antibiotic (e.g., 100 μg/mL ampicillin) and incubate for 24 hours at 37°C.
- Inoculate a single colony into 10 mL of LB medium with antibiotic and grow overnight at 37°C on a rotary shaker at 200 rpm.
- The following day, dilute the culture to ~10⁷ cells/mL and regrow without antibiotics to the early exponential phase (OD₆₀₀ ≈ 0.2) [34].
2. Cell Immobilization in Hydrogel:
- Harvest the bacterial cells via centrifugation.
- Mix the cell pellet 1:1 (v/v) with a 2.5% (w/v) sodium alginate solution.
- Load the cell-alginate mixture into custom cellulose tubes (e.g., 6 mm diameter).
- Immerse the tubes in a 0.25 M calcium chloride (CaCl₂) solution for 20 minutes to initiate polymerization and form stable calcium alginate tablets.
- Remove the alginate tubes and cut them into 3 mm tablets using a customized holder [34].
3. Biosensor Activation and Measurement:
- Expose the bacterial tablets to the air sample or headspace from the crop of interest (e.g., in a storage container).
- Monitor the bioluminescent response using a multi-mode plate reader or an imaging system like the IVIS Lumina LT.
- The induction of bioluminescence above a baseline threshold indicates the presence of target VOCs, signaling potential spoilage [34]. Lower temperatures (+4°C) have been shown to enhance sensor sensitivity and prolong bacterial viability, making this ideal for cold storage monitoring [34].
The logical relationship between the sensor components and the signal output is shown below.
Integration and Future Perspectives
The true potential of monitoring plant physiochemical signals is realized when sap analysis and biosensor data are integrated into a comprehensive decision-support system. Future trends point towards the development of miniaturized, non-invasive, and intelligent sensors with long-term stability [31]. The integration of Artificial Intelligence (AI) and machine learning is revolutionizing data analytics, enabling stress pattern recognition, event forecasting, and the design of targeted interventions [35] [16]. These systems, when validated at scale, can serve as replicable models for precision agriculture, allowing for closed-loop feedback systems that automatically optimize irrigation and nutrient delivery [16].
Despite the promise, challenges remain in the widespread adoption of these technologies, particularly for electrochemical biosensors. Key hurdles include ensuring sensor stability and longevity in complex real-world matrices, mitigating fouling and interference, establishing standardized calibration protocols, and achieving user-friendly design for end-users [31] [16]. Overcoming these barriers through continued innovation in materials, manufacturing, and stakeholder engagement is essential to advancing from laboratory prototypes to robust field-deployable tools that will ultimately enhance global food security.
The growing pressure on global agricultural systems necessitates advanced monitoring tools to ensure soil health and food safety. Soil contamination from pesticides, heavy metals, and emerging pollutants poses a significant threat to ecosystems, crop quality, and human health. These contaminants originate from various agricultural and industrial practices, leading to persistent environmental accumulation and entry into the food chain [36] [37]. Within this context, electrochemical biosensors have emerged as powerful analytical tools, offering rapid, sensitive, and field-deployable solutions for precision agriculture [16]. These devices combine a biological recognition element with an electrochemical transducer, converting a biological interaction into a quantifiable electrical signal [38]. This technical guide provides an in-depth examination of electrochemical biosensing technologies for soil contaminant detection, detailing fundamental principles, current applications, experimental protocols, and future directions to support research and development in agricultural science.
Fundamental Principles of Electrochemical Biosensors
Electrochemical biosensors are characterized by their core components: a biorecognition element and an electrochemical transducer. The biorecognition element, which can be an enzyme, antibody, DNA strand, or aptamer, provides specificity by interacting with the target analyte [16] [38]. The transducer then converts this biological interaction into a measurable electrical signal—such as current, potential, or impedance—that is proportional to the analyte concentration [38].
The operational principle can be further categorized into several detection mechanisms:
- Voltammetric Techniques: These measure current as a function of applied potential, providing high sensitivity for electroactive species. Common methods include square-wave voltammetry (SWV) and differential pulse voltammetry (DPV).
- Amperometric Techniques: These monitor current at a fixed potential, which changes upon the biochemical reaction at the electrode surface.
- Potentiometric Techniques: These measure the potential difference between working and reference electrodes under conditions of zero current.
- Impedimetric Techniques: These track changes in the impedance of the electrode interface, often used for label-free detection [12] [38].
A critical advancement in this field is the integration of nanomaterials—such as metal nanoparticles, carbon nanotubes, and graphene—into sensor designs. These materials enhance the electrode's effective surface area, improve electron transfer kinetics, and can be functionalized to increase biomolecule immobilization, thereby boosting the sensor's sensitivity, selectivity, and stability [12] [3].
The following diagram illustrates the fundamental components and operational workflow of a typical electrochemical biosensor for soil contaminant detection.
Electrochemical Biosensors for Major Soil Contaminants
Pesticide Detection
Pesticides, including organophosphates, carbamates, and triazines, are extensively used in agriculture but pose serious risks due to their environmental persistence and potential toxicity to non-target organisms [36] [37]. Electrochemical biosensors for pesticide detection often employ enzymes such as acetylcholinesterase (AChE), tyrosinase, or alkaline phosphatase as biorecognition elements. The detection mechanism typically relies on the inhibition of these enzymes by the pesticide, leading to a measurable decrease in electrochemical signal [37].
Recent innovations focus on enhancing sensor performance through nanomaterial integration. For instance, nanostructured metal oxides and carbon-based materials significantly lower the detection limit and improve stability in complex soil matrices [12]. A notable development is the use of non-enzymatic nanoceria tags and molecularly imprinted polymers (MIPs) that mimic biological recognition, offering robust and cost-effective alternatives for organophosphate pesticide detection [16] [37].
Heavy Metal Detection
Heavy metals such as cadmium (Cd), lead (Pb), mercury (Hg), and arsenic (As) are toxic, non-biodegradable, and tend to bioaccumulate, making them a priority for environmental monitoring [37] [39]. Electrochemical biosensors for heavy metals frequently utilize DNAzymes (catalytic DNA), aptamers (single-stranded DNA/RNA), or whole cells as biological elements. These probes undergo conformational changes or cleavage upon binding specific metal ions, generating a distinct electrochemical signal [38] [39].
For example, ssDNA-based sensors have been developed where the interaction with heavy metals causes guanine oxidation signals to diminish, enabling highly sensitive detection. Nanomaterial-enhanced platforms, such as those using gold-decorated polymer nanofibers or ZnO nanoparticles, have demonstrated exceptional performance for detecting Cu²⁺, Cd²⁺, and Hg²⁺ ions in environmental samples, often achieving detection limits far below regulatory thresholds [38] [40].
Detection of Emerging Pollutants
Emerging contaminants (ECs) encompass a diverse range of substances, including pharmaceuticals, personal care products, endocrine disruptors, and industrial chemicals, whose environmental impact is not fully understood [36]. These compounds often coexist in soil, leading to potential synergistic toxic effects. Electrochemical biosensors designed for ECs leverage affinity-based recognition elements like antibodies and aptamers for high-specificity detection [36] [12].
Significant progress has been made in multiplexed sensor platforms that can simultaneously detect several ECs. The integration with molecularly imprinted polymers (MIPs) creates artificial recognition sites complementary to the target molecule, enhancing sensor robustness and longevity in field conditions [12] [39].
Table 1: Performance Metrics of Electrochemical Biosensors for Soil Contaminants
| Target Contaminant | Biorecognition Element | Detection Technique | Linear Range | Limit of Detection (LOD) | Reference |
|---|---|---|---|---|---|
| Pesticides (e.g., Organophosphates) | Acetylcholinesterase Enzyme | Amperometry | Varies by compound | Nanomolar to picomolar range | [16] [37] |
| Cadmium (Cd²⁺) | ssDNA / Aptamer | Square-Wave Voltammetry (SWV) | Not specified | ~0.071 µg/L (for analogous leucine detection) | [38] [40] |
| Lead (Pb²⁺) | DNAzyme | Voltammetry | Not specified | Enhanced sensitivity with nanomaterials | [38] |
| Mercury (Hg²⁺) | Aptamer / Gold-nanoparticle composite | Differential Pulse Voltammetry (DPV) | Not specified | Ultralow concentrations (specific values vary) | [16] [38] |
| Emerging Contaminants (e.g., Pharmaceuticals) | Antibody / Molecularly Imprinted Polymer (MIP) | Impedimetry / Voltammetry | Compound-dependent | Nanomolar range | [36] [12] |
Experimental Protocols for Sensor Development and Application
Protocol 1: Development of a Carbon Paste Electrode (CPE) Modified with ssDNA for Contaminant Detection
This protocol outlines the procedure for creating a robust biosensor platform, adaptable for detecting heavy metals or organic pollutants that interact with DNA [40].
Materials:
- Graphite powder and mineral oil for carbon paste preparation.
- Single-stranded DNA (ssDNA) with a high guanine content.
- Buffer solutions: Acetate buffer (0.1 M, pH 5.0) and phosphate buffer saline (PBS, 0.1 M, pH 7.4).
- Target analyte standard (e.g., cadmium standard solution).
- Electrochemical workstation with three-electrode setup.
Procedure:
- Electrode Fabrication: Mix graphite powder and mineral oil in a 70:30 ratio by weight to form a homogeneous carbon paste. Pack the paste firmly into a Teflon electrode body with an electrical contact.
- ssDNA Immobilization: Deposit 5–10 µL of a thermally denatured ssDNA solution (e.g., 100 µg/mL) onto the CPE surface. Allow it to dry at room temperature to form a stable film.
- Interaction with Analyte: Immerse the ssDNA-modified CPE in a solution containing the target contaminant (e.g., soil extract). Incubate for a fixed period (e.g., 5–15 minutes) to allow binding, which may cause guanine oxidation signals to decrease.
- Electrochemical Measurement: Perform Square-Wave Voltammetry (SWV) in a clean acetate buffer solution. Apply a potential from +0.5 V to +1.0 V (vs. Ag/AgCl) to monitor the guanine oxidation peak at approximately +0.86 V.
- Data Analysis: Quantify the target concentration by measuring the decrease in the guanine peak height relative to a calibration curve constructed from standard solutions.
Protocol 2: Sample Preparation and Analysis of Soil Extracts
Accurate detection in real soil matrices requires careful sample preparation to minimize interference.
Materials:
- Soil sample.
- Extraction solvent (e.g., 0.1 M CH₃COONH₄, pH 7.0, or 2 M KCl for free amino acids).
- Centrifuge and filtration units (0.45 µm membrane).
- pH meter and standard buffer solutions.
Procedure:
- Soil Extraction: Air-dry and sieve the soil sample through a 2-mm mesh. Weigh 10 g of soil and mix with 20 mL of extraction solvent. Shake the mixture vigorously for 30 minutes.
- Clarification: Centrifuge the soil suspension at 5000 rpm for 15 minutes. Filter the supernatant through a 0.45 µm membrane filter to obtain a clear extract.
- pH Adjustment: Adjust the pH of the soil extract to a value compatible with the biosensor (typically between 5.0 and 8.0). Avoid drastic pH changes that can alter contaminant bioavailability.
- Analysis with Biosensor: Dilute the extract if necessary and analyze using the calibrated electrochemical biosensor following the specific measurement protocol (e.g., SWV, DPV).
- Validation: Validate the method's accuracy by spiking the soil sample with a known concentration of the target analyte and calculating the recovery rate.
The following workflow diagram summarizes the key steps in soil contaminant analysis, from sensor preparation to final detection.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for Electrochemical Biosensor Development
| Reagent/Material | Function | Example Application |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, portable electrode platforms for field-deployable sensing. | Base transducer for on-site detection of pesticides and heavy metals [38]. |
| Nanomaterials (CNTs, Graphene, Metal NPs) | Enhance electrode conductivity, surface area, and biomolecule immobilization. | Signal amplification in sensors for pathogens and mycotoxins in crops [3]. |
| Specific Bioreceptors (Aptamers, DNAzymes) | Provide high-affinity molecular recognition for target analytes. | Selective binding to heavy metal ions like Pb²⁺ and Cd²⁺ [38] [40]. |
| Enzymes (AChE, Tyrosinase) | Catalyze reactions with specific substrates; inhibition signals pesticide presence. | Core recognition element in organophosphate and carbamate pesticide biosensors [16] [37]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic, stable polymers with tailor-made cavities for specific analyte binding. | Detection of emerging contaminants (e.g., pharmaceuticals) in complex matrices [12] [39]. |
| Buffer Salts (PBS, Acetate Buffer) | Maintain stable pH and ionic strength during bio-recognition and electrochemical measurement. | Essential medium for all electrochemical biosensing experiments [40]. |
The field of electrochemical biosensing for soil monitoring is rapidly evolving, driven by interdisciplinary innovations. Key future research directions include the integration of artificial intelligence (AI) and machine learning for advanced data interpretation, which can deconvolute complex signals from soil matrices and improve predictive capabilities [16] [3]. The development of biodegradable and sustainable sensor materials will also reduce the environmental footprint of these monitoring tools [3].
Furthermore, the convergence of biosensors with Internet of Things (IoT) platforms is a critical step towards smart agriculture. This integration enables the creation of wireless sensor networks for real-time, continuous soil monitoring, providing actionable data for precision farming and early warning systems [16] [39]. Overcoming existing challenges related to sensor stability, standardization, and user-friendly design will accelerate the transition of these technologies from laboratory prototypes to indispensable tools for environmental stewardship and sustainable agriculture [16] [12].
In conclusion, electrochemical biosensors represent a paradigm shift in soil contaminant detection. Their high sensitivity, portability, and potential for real-time analysis make them powerful tools for researchers and agricultural professionals dedicated to safeguarding soil health, ensuring food safety, and promoting sustainable agricultural practices.
Electrochemical biosensors are emerging as transformative tools in precision agriculture, enabling the direct, on-site detection of pathogens, nutrients, and environmental stressors. These analytical devices combine a biological recognition element with an electrochemical transducer, converting a biological event into a quantifiable electrical signal. The integration of these biosensors with Internet of Things (IoT) architectures, Artificial Intelligence (AI) for data processing, and miniaturized Lab-on-a-Chip (LOC) platforms is creating a new paradigm for intelligent, data-driven farm management. This integration facilitates a closed-loop system where real-time sensor data informs automated decisions, optimizing resource use and enhancing crop protection within the framework of Agriculture 4.0 and 5.0 [41].
Despite their promise, a significant gap exists between laboratory development and field deployment. A systematic review highlights that a vast majority of electrochemical biosensor studies rely on artificially spiked samples rather than real-world validation, with only 1 out of 77 studies conducting direct testing on naturally contaminated matrices [7]. This underscores the critical need for robust integration frameworks that can handle the complexities of agricultural environments. This whitepaper provides a technical guide to the core components, experimental methodologies, and implementation protocols for effectively merging electrochemical biosensing with smart agriculture systems.
Core Components of an Integrated Smart Sensing System
Advanced Electrochemical Biosensors
Electrochemical biosensors form the frontline data acquisition layer. Their performance is critically determined by the interface engineering of the electrode surface.
- Nanomaterial-Enhanced Electrodes: The integration of nanomaterials like multi-walled carbon nanotubes (MWCNTs), graphene, and metal nanoparticles has been pivotal in enhancing signal amplification, conductivity, and the effective surface area of electrodes. These materials lower the limit of detection (LOD) by facilitating electron transfer and increasing the loading capacity of biorecognition elements [7] [3].
- Biorecognition Elements: The specificity of biosensors is derived from biological receptors. Common elements include:
- Antibodies: Valued for high specificity, used in immunosensors for pathogen detection [7].
- Aptamers: Single-stranded DNA or RNA molecules selected for high affinity to targets; offer advantages in stability and synthesis over antibodies [3] [42].
- Nucleic Acid Probes: Used for detecting pathogen DNA or RNA, often integrated with isothermal amplification techniques for field use [22].
- Bacteriophages: Genetically engineered phages are used as stable and specific recognition agents for bacteria like E. coli [7].
- Electrochemical Techniques: Different techniques are employed based on the target analyte and required sensitivity, as detailed in Table 1.
Table 1: Common Electrochemical Detection Techniques and Their Agricultural Applications
| Technique | Measurement Principle | Key Applications in Agriculture | Advantages |
|---|---|---|---|
| Amperometry | Measures current from redox reactions at a constant potential. | Glucose monitoring, detection of metabolites, pathogen presence [43]. | High sensitivity, compatible with miniaturization. |
| Voltammetry (e.g., DPV, SWV) | Measures current while scanning a range of applied potentials. | Detection of heavy metals, mycotoxins, pathogen DNA [43] [22]. | Low-noise signal, capable of multi-analyte detection. |
| Electrochemical Impedance Spectroscopy (EIS) | Measures impedance (resistance & capacitance) across a frequency range. | Label-free detection of pathogen binding, soil analysis [43]. | Label-free, monitors binding events in real-time. |
| Potentiometry | Measures potential difference between electrodes at near-zero current. | Soil pH monitoring, ion-selective detection (K+, NO3-) [43]. | Simple, low-power, resistant to interfacial interference. |
Internet of Things (IoT) and Connectivity
The IoT framework enables the transformation of standalone biosensors into a networked system for real-time monitoring. It comprises several layers:
- Sensor Node: Contains the electrochemical biosensor, a microcontroller (e.g., ESP32), and signal conditioning circuits. The node performs the initial analog-to-digital conversion of the electrochemical signal [44].
- Communication Protocols: Wireless protocols like LoRaWAN, Zigbee, or cellular networks (4G/5G) transmit data from field-deployed sensor nodes to a central gateway [44] [41].
- Cloud/Edge Platform: Data is aggregated and stored in cloud platforms or processed at the edge to reduce latency. This layer supports data visualization, historical analysis, and system control via farmer-facing mobile applications [44].
Artificial Intelligence and Machine Learning
AI, particularly machine learning (ML) and deep learning (DL), is infused throughout the system to overcome the limitations of traditional analysis and handle complex, multivariate data [42].
- Signal Processing and Optimization: ML algorithms such as Support Vector Machines (SVMs) and Artificial Neural Networks (ANNs) are employed to process raw electrochemical data, filter noise, and correct for baseline drift and matrix effects from complex samples like plant sap or soil extracts [45] [42].
- Predictive Analytics and Diagnostics: AI models analyze sensor data alongside historical and environmental data to predict disease outbreaks, nutrient deficiencies, and optimal harvest times. This enables predictive analytics for proactive farm management [44] [41].
- Sensor Design and Optimization: At the design stage, AI-driven models assist in predicting the binding affinity of aptamers, optimizing electrode material compositions, and tuning electrochemical parameters, thereby accelerating the development of high-performance sensors [42].
The following diagram illustrates the architecture and information flow of an integrated system.
The Scientist's Toolkit: Key Research Reagent Solutions
The development and operation of integrated electrochemical biosensors rely on a suite of essential reagents and materials. The following table details these key components and their functions in a typical research or deployment scenario.
Table 2: Essential Research Reagents and Materials for Integrated Agricultural Biosensing
| Category | Item | Primary Function in Experiments |
|---|---|---|
| Biorecognition Elements | Specific aptamers (e.g., for S. sclerotiorum) | Target capture and sensor specificity; selected via SELEX or AI-predicted binding affinity [3] [42]. |
| Monoclonal/polyclonal antibodies (e.g., against aflatoxin B1) | Immunosensing; provide high affinity and specificity to pathogens or toxins [7]. | |
| DNA probes (e.g., for CaMV 35S promoter in GMO detection) | Nucleic acid hybridization for detecting pathogen DNA or genetically modified elements [22]. | |
| Nanomaterials for Electrode Modification | Multi-walled Carbon Nanotubes (MWCNTs) | Enhance electrode conductivity and surface area; improve electron transfer kinetics and signal-to-noise ratio [7] [3]. |
| Graphene oxide & Metal Nanoparticles (e.g., Au, Ag NPs) | Signal amplification; often used as carriers for immobilizing biorecognition elements or as redox labels [3]. | |
| Signal Generation & Amplification | Redox markers (e.g., Methylene Blue, [Fe(CN)₆]³⁻/⁴⁻) | Produce measurable electrochemical current; change in signal indicates binding event [7] [22]. |
| Enzyme labels (e.g., Horseradish Peroxidase - HRP) | Catalyze substrate reaction for amplified signal output in enzyme-linked assays [7]. | |
| Sample Preparation & Assay | Buffer solutions (e.g., PBS, Tris-HCl) | Maintain optimal pH and ionic strength for biomolecular interactions and electrochemical stability [43]. |
| Blocking agents (e.g., BSA, casein) | Minimize non-specific binding on the sensor surface, reducing false-positive signals [7]. |
Experimental Protocols for System Development and Validation
Protocol: Development of a Nanomaterial-Modified Biosensor
Aim: To fabricate and characterize a nanomaterial-enhanced electrochemical biosensor for the detection of a specific plant pathogen (e.g., Phakopsora pachyrhizi, soybean rust).
Materials:
- Glassy carbon electrode (GCE) or screen-printed carbon electrode (SPCE)
- Functionalized multi-walled carbon nanotubes (f-MWCNTs)
- Gold nanoparticle (AuNP) colloid
- Pathogen-specific aptamer solution (e.g., 1 µM in Tris-EDTA buffer)
- Methylene blue redox probe
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
Method:
- Electrode Pretreatment: Polish the GCE with alumina slurry (0.05 µm) and rinse thoroughly with deionized water. For SPCEs, this step may be omitted.
- Nanomaterial Modification:
- Prepare a dispersion of f-MWCNTs (1 mg/mL) in a suitable solvent (e.g., DMF).
- Drop-cast 5-10 µL of the dispersion onto the electrode surface and allow it to dry under an infrared lamp.
- Subsequently, drop-cast 5 µL of the AuNP colloid to form an MWCNT/AuNP hybrid layer.
- Aptamer Immobilization:
- Incubate the modified electrode with the aptamer solution for 60 minutes at room temperature.
- Rinse gently with PBS to remove unbound aptamers.
- Blocking: Treat the electrode with 1% BSA solution for 30 minutes to block any remaining active sites and prevent non-specific adsorption.
- Electrochemical Characterization:
- Use Cyclic Voltammetry (CV) and Electrochemical Impedance Spectroscopy (EIS) in a 5 mM [Fe(CN)₆]³⁻/⁴⁻ solution to monitor the modification process after each step. A successful modification is indicated by a decrease in peak current (CV) and an increase in charge transfer resistance (EIS) after aptamer immobilization.
- Detection and Calibration:
- Incubate the biosensor with samples containing varying concentrations of the target pathogen.
- Perform Differential Pulse Voltammetry (DPV) in a solution containing methylene blue.
- Record the change in peak current, which correlates with the target concentration. Plot a calibration curve of current vs. log(concentration) to determine the LOD and linear range.
Protocol: Integration with an IoT Node and Data Acquisition
Aim: To interface the biosensor with a microcontroller for wireless data transmission.
Materials:
- Fabricated biosensor (SPCE recommended)
- Potentiostat (miniature or integrated shield, e.g., with ESP32)
- Microcontroller (e.g., ESP32)
- Power source (battery)
- IoT communication module (e.g., LoRa module)
Method:
- Hardware Interfacing: Connect the potentiostat shield to the microcontroller. Connect the working, reference, and counter terminals of the biosensor to the corresponding terminals on the potentiostat.
- Firmware Programming: Program the microcontroller to:
- Execute the DPV protocol with predefined parameters (potential window, pulse amplitude, step potential).
- Read the measured current data from the potentiostat via I2C or SPI communication.
- Timestamp the acquired data.
- Data Transmission: Package the sensor data (sensor ID, timestamp, current value) into a JSON format and transmit it to a cloud server via the LoRaWAN network at set intervals.
- Cloud Data Ingestion: Configure the cloud platform (e.g., AWS IoT Core, The Things Stack) to receive and decode the data packets, storing them in a database for further analysis and visualization.
Protocol: AI-Assisted Data Analysis for Pathogen Identification
Aim: To employ a machine learning model to classify and quantify pathogens from multiplexed sensor data.
Materials:
- Dataset of DPV responses from sensors exposed to different pathogens and concentrations.
- Python environment with scikit-learn and TensorFlow/PyTorch libraries.
Method:
- Data Preprocessing:
- Collect raw DPV current-potential data.
- Perform baseline correction (e.g., using asymmetric least squares smoothing).
- Normalize the current values to a 0-1 scale.
- Split the dataset into training (70%), validation (15%), and test (15%) sets.
- Model Selection and Training:
- For classification (identifying the pathogen type), train a Convolutional Neural Network (CNN) on the preprocessed DPV voltammograms, treating them as 1D signals.
- For regression (determining the concentration), train a Random Forest regressor or a Support Vector Regressor (SVR) using features extracted from the DPV peaks (e.g., peak height, peak area, peak potential).
- Model Evaluation: Validate the model performance on the test set using metrics such as accuracy, precision, recall, and F1-score for classification, and Mean Absolute Error (MAE) and R² score for regression.
- Deployment: Deploy the trained model on the cloud or edge server to analyze incoming sensor data in real-time, automatically generating diagnostic reports and alerts.
Future Perspectives and Challenges
The convergence of electrochemical biosensors with IoT and AI is poised to redefine precision agriculture. Key future directions include:
- Tackling the Real-World Validation Gap: Future research must prioritize validation using naturally contaminated samples in field conditions to bridge the gap between laboratory performance and on-farm reliability [7].
- Advanced Material Science: The development of biodegradable sensor substrates and self-healing conductive materials will enhance environmental sustainability and device longevity [3].
- Sophisticated AI Integration: AI will evolve from a data analysis tool to a core component of sensor design and adaptive system control. This includes using AI for dynamic calibration to correct for sensor drift and environmental interference [42].
- Standardization and Regulation: Establishing standardized validation protocols and achieving regulatory alignment with bodies like the FAO and FDA is crucial for the widespread adoption and commercial success of these integrated systems [7].
The primary challenges remain the high initial implementation cost, the need for technical expertise, data security concerns, and ensuring connectivity in remote agricultural areas [46] [41]. Overcoming these hurdles through collaborative efforts between researchers, industry stakeholders, and policymakers will be essential to fully realize the potential of integrated smart sensors in building a more resilient and productive agricultural system.
Overcoming Field Deployment Challenges: Optimization Strategies and Nanomaterial Engineering
Addressing Matrix Interference from Complex Plant and Soil Samples
The accurate detection of specific analytes in plant and soil samples is fundamentally challenged by matrix effects, where the complex composition of the sample itself interferes with the measurement. For electrochemical biosensors, these effects can manifest as signal suppression or enhancement, leading to inaccurate quantification and potentially compromising agricultural research outcomes. Soil is an exceptionally difficult medium due to its spatial heterogeneity, opacity, and varied physical and chemical properties that change with depth, time, and microbial activity [47]. Plant tissues present similar challenges with their complex mixtures of organic compounds, pigments, and ionic content. Understanding and mitigating these interferences is therefore a critical prerequisite for deploying reliable electrochemical biosensors in an agricultural context.
This guide synthesizes current strategies to characterize, compensate for, and minimize matrix effects, providing a technical roadmap for researchers developing and applying these biosensing platforms.
Matrix effects arise from the combined influence of all sample components other than the target analyte. These can be categorized as follows:
- Physical Effects: Soil is an opaque, heterogeneous mixture, which limits the use of optical techniques and creates variability between sub-samples [47]. This physical complexity can hinder analyte access to the biosensor's recognition element.
- Chemical Effects: The soil and plant matrices contain numerous compounds that can interfere. These include inorganic ions that alter ionic strength, organic matter (e.g., humic acids), and secondary metabolites that may nonspecifically bind to sensor surfaces or compete for binding sites [48] [49].
- Biological Effects: The presence of billions of microbes per gram of soil can metabolize the target analyte or the biosensor's components, affecting the signal over time [47].
In electrochemical biosensors, these interferences can alter electron transfer kinetics at the electrode surface or cause fouling, reducing sensitivity and selectivity. A key advantage of electrochemical sensing is its relative robustness to optical interferences like turbidity and color, which plague fluorescence- and Raman-based methods [50].
Strategic Framework for Mitigating Matrix Effects
A systematic approach to managing matrix effects involves first evaluating their severity and then selecting an appropriate combination of the following strategies.
Sample Preparation and Clean-Up
The primary goal of sample preparation is to reduce matrix complexity before analysis.
- Liquid Extraction: A common first step is to sieve soil through a 2-mm sieve and incubate it in a liquid medium, with the supernatant extracted for analysis [47]. However, this method may not transfer all microbes or analytes.
- Enzymatic Liquefaction: For viscous samples, enzymatic methods can be used. One protocol involves adding hydrogen peroxide for 1 minute to mechanically disrupt the sample matrix through bubble production without harsh chemicals or instrumentation [51].
- Commercial Kits: Numerous DNA/RNA extraction kits are tailored for soil (e.g., FastDNA SPIN kits, DNeasy Power kits, ZymoBIOMICS DNA kits). It is critical to select a kit appropriate for the specific soil type, as each has its own biases and recovery efficiencies [47].
Sensor Design and Interface Engineering
Innovations at the biosensor interface can inherently improve resistance to interference.
- Stable Bioreceptors: Aptamers (single-stranded DNA or RNA oligonucleotides) are noted for their stability compared to enzymes or antibodies and are particularly promising for field-use biosensors in complex environments [47] [49].
- Nanostructured Electrodes: The use of nanomaterials like gold nanoparticles (AuNPs), graphene, and carbon nanotubes can enhance sensitivity and selectivity. Their high surface-to-volume ratio maximizes active sites, and their conductive pathways accelerate electron transfer [15] [50].
- Multiplexed and Calibrated Sensing: A multichannel sensor design allows for in-situ calibration and includes negative controls. This enables statistical validation of results and helps correct for sample-to-sample matrix variability [48].
Signal Correction and Calibration Methods
When matrix effects cannot be fully eliminated, computational and calibration techniques are essential.
- Matrix-Matched Calibration: This involves preparing calibration standards in a blank matrix that is chemically similar to the sample (e.g., a clean soil extract). This helps ensure that the calibration curve experiences similar matrix effects as the sample [52] [53].
- Standard Addition: The sample is spiked with known concentrations of the target analyte, and the signal change is measured. This method accounts for the matrix effect directly within the sample itself.
- Internal Standards: Using an isotope-labeled internal standard is considered a highly effective approach. The standard is added to the sample at the beginning of analysis, and its recovery is used to correct for analyte loss and signal suppression or enhancement during sample preparation and analysis [52].
Table 1: Summary of Matrix Effect Compensation Strategies
| Strategy | Principle | Best Used When | Limitations |
|---|---|---|---|
| Matrix-Matched Calibration | Calibrants and samples have matching background matrix [52]. | A blank matrix is readily available. | Finding a truly blank matrix can be difficult. |
| Standard Addition | Spike and measure recovery directly in the sample [53]. | Sample volume is sufficient and matrix is highly variable. | More labor-intensive; requires multiple analyses per sample. |
| Isotope-Labeled Internal Standard | A chemically identical standard corrects for losses and ME [52]. | High accuracy is required; for quantitative LC-MS. | Expensive; may not be available for all analytes. |
| Slope Ratio Analysis | Compares calibration slope in solvent vs. matrix [52]. | For semi-quantitative screening of ME magnitude. | Does not fully correct data; used for evaluation. |
Detailed Experimental Protocols
Protocol 1: Post-Column Infusion for Qualitative ME Assessment
This protocol helps identify chromatographic regions affected by ion suppression or enhancement in LC-MS-based methods [52].
- Setup: Integrate a T-piece between the HPLC column outlet and the mass spectrometer inlet.
- Infusion: Continuously infuse a standard solution of the target analyte (at a concentration within the analytical range) through the T-piece at a constant flow rate.
- Injection: Inject a blank, prepared sample extract (e.g., a soil or plant extract) onto the HPLC column. Use a standard gradient elution method.
- Detection: The mass spectrometer will display a steady baseline from the infused analyte, which will dip or rise when matrix components from the blank extract co-elute and cause ion suppression or enhancement.
- Analysis: The resulting chromatogram identifies retention time windows where matrix effects occur, guiding adjustments to the chromatography or sample clean-up.
Protocol 2: Paper-Based Biosensor for Pyocyanin in Sputum (Adaptable for Soil Extracts)
This protocol demonstrates a sensor design that mitigates matrix effects for complex samples [51].
- Sensor Fabrication:
- Substrate: Cut Whatman #41 filter paper into 2 x 2 cm squares.
- Competing Element: Coat the paper with 10 µL of an albumin-antigen conjugate (PC1-BSA, 1.52 µM) and let it dry.
- Reservoir: Prepare a separate piece of PSS-infused filter paper as a reservoir containing 20 nm gold nanoparticles modified with a specific monoclonal antibody (mAb122).
- Sample Preparation:
- Liquefy the complex sample (e.g., soil slurry or plant homogenate) using a mild enzymatic method or a 1-minute H₂O₂ treatment [51].
- Centrifuge and collect the supernatant.
- Assay Procedure:
- Apply the liquefied sample to the paper substrate.
- Press the Ab-AuNP reservoir against the substrate and incubate for 5 minutes. During this time, the AuNPs interact with both free PYO from the sample and the paper-bound PC1-BSA in a competitive manner.
- Wash the substrate. The color intensity of the spot is inversely proportional to the analyte concentration in the sample.
- Key Advantage: This competitive immunoassay format on a paper platform has been shown to reduce matrix effects compared to traditional ELISA, yielding lower relative standard deviations in complex samples [51].
The logical workflow for selecting an appropriate mitigation strategy based on the nature of the interference and analytical requirements is summarized below.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents and Materials for Mitigating Matrix Effects
| Reagent/Material | Function | Example Application |
|---|---|---|
| Aptamers | Single-stranded DNA/RNA oligonucleotides serving as synthetic, stable bioreceptors [47] [49]. | Specific detection of heavy metals or organic pollutants; more stable than antibodies for field-use sensors. |
| Gold Nanoparticles (AuNPs) | Nanomaterials for signal amplification and electron transfer enhancement [51] [50]. | Used in paper-based immunosensors and for modifying electrodes to lower detection limits. |
| Isotope-Labeled Internal Standards | Chemically identical standards for accurate quantification, correcting for analyte loss and ME [52]. | Essential for quantitative LC-MS analysis of contaminants in plant/soil extracts. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymers with cavities complementary to the target analyte, offering selective extraction [52]. | Solid-phase extraction clean-up to remove interfering compounds from complex samples. |
| Commercial DNA/RNA Kits | Standardized reagents for efficient nucleic acid extraction from complex matrices [47]. | Soil microbiome characterization (e.g., MP Biomedicals FastDNA SPIN kit, Qiagen DNeasy Power kit). |
| Enzymatic Liquefaction Reagents | Enzymes (e.g., lysozyme) and H₂O₂ to disrupt complex, viscous samples [51]. | Pre-treatment of soil slurries or plant root samples to release analytes and reduce viscosity. |
Addressing matrix interference is not a one-time task but an integral part of the biosensor development and validation process. A combination of thoughtful sample preparation, robust sensor design, and appropriate calibration strategies is required to generate reliable data from complex plant and soil samples.
Future advancements will likely integrate machine learning to analyze complex datasets from multiplexed sensors and decipher signal patterns obscured by matrix interference [47]. Furthermore, the development of AI-assisted data interpretation and the integration of biosensors with agricultural IoT networks will pave the way for real-time, field-deployable disease and contaminant monitoring, ultimately contributing to sustainable agriculture and food security [15]. By systematically applying the principles and protocols outlined in this guide, researchers can significantly enhance the accuracy and utility of electrochemical biosensors in agricultural research.
The integration of nanotechnology into electrochemical biosensor design has revolutionized analytical capabilities in agricultural research, enabling precise, on-site detection of pathogens, pesticides, and environmental contaminants [54]. These transformations are largely driven by innovative nanomaterial combinations, particularly gold nanoparticles (AuNPs) and graphene-based materials, which collectively enhance sensor sensitivity, selectivity, and stability [55] [56]. This technical guide examines the fundamental mechanisms, fabrication methodologies, and practical applications of AuNP-graphene hybrid systems for signal amplification in electrochemical biosensors, with specific emphasis on agricultural monitoring applications. The unique synergistic effects between these nanomaterials address critical challenges in detecting low-concentration analytes within complex agricultural matrices, providing researchers with powerful tools for crop disease management, food safety assurance, and environmental protection [57] [3].
Fundamental Principles of Signal Amplification
Synergistic Effects of AuNPs and Graphene
The enhanced performance of AuNP-graphene nanocomposites in electrochemical biosensors stems from synergistic effects that amplify detection signals through multiple mechanisms. These hybrid materials create an optimal environment for biomolecule immobilization and electron transfer, significantly improving sensor performance compared to either material used independently [58] [56].
Table 1: Signal Amplification Mechanisms of AuNP-Graphene Nanocomposites
| Amplification Mechanism | Contribution of Graphene | Contribution of AuNPs | Synergistic Effect |
|---|---|---|---|
| Enhanced Surface Area | Large specific surface area (2630 m²/g) provides extensive immobilization sites [56] | Nanoparticles further increase effective surface area | Maximized analyte capture capacity; higher receptor density |
| Improved Electron Transfer | High intrinsic charge carrier mobility (200,000 cm²/V·s) [56] | Facilitate electron tunneling; act as conductive bridges | Rapid electron transport throughout electrode matrix |
| Catalytic Activity | Limited intrinsic catalysis but excellent support material | Intrinsic peroxidase-like and catalytic properties [55] | Enhanced reaction kinetics for electrochemical processes |
| Biomolecule Immobilization | π-π stacking, hydrophobic interactions for aromatic molecules [57] | Thiol-Au covalent bonding for biomolecules [58] | Versatile attachment strategies for diverse recognition elements |
Graphene's two-dimensional honeycomb lattice provides exceptional electrical conductivity, substantial specific surface area (2630 m²/g), and remarkable mechanical flexibility, establishing an ideal foundation for biosensor development [56]. When incorporated into electrochemical sensors, graphene sheets form continuous conductive networks that facilitate rapid electron transfer between immobilized biomolecules and electrode surfaces [57]. The material's high surface-to-volume ratio enables dense loading of recognition elements such as enzymes, antibodies, and aptamers, significantly increasing the probability of analyte capture [56].
AuNPs contribute complementary advantages, including excellent biocompatibility, high surface energy, and unique optical and electrical properties [58]. Their capacity for strong thiol-Au interactions facilitates stable immobilization of biomolecules while preserving biological activity [59]. More importantly, AuNPs exhibit intrinsic catalytic activity toward numerous electrochemical reactions, enabling direct signal amplification without additional reagent labeling [55]. When dispersed on graphene surfaces, AuNPs prevent graphene sheet restacking through spacer effects, maintaining the accessible surface area while introducing additional catalytic sites [60].
The synergistic coupling between these nanomaterials creates composite structures with enhanced charge transfer capabilities, increased active surface area, and improved biomolecule stability [58]. Electrons can travel efficiently through the continuous graphene network while AuNPs act as nanoscale conductive bridges, minimizing electron transfer resistance and reducing activation energy barriers for electrochemical reactions [60]. This multidimensional amplification strategy enables detection of target analytes at exceptionally low concentrations, meeting the demanding requirements of agricultural monitoring applications where pathogens and contaminants may be present at trace levels [3].
Charge Transfer Mechanisms in Hybrid Nanocomposites
The exceptional electrical properties of AuNP-graphene nanocomposites originate from complementary charge transfer mechanisms that operate simultaneously within the hybrid structure. Graphene's sp²-hybridized carbon lattice provides a continuous pathway for electron delocalization, enabling rapid charge carrier mobility exceeding 200,000 cm²/V·s in pristine samples [56]. This intrinsic conductivity establishes the foundation for efficient electron transport throughout the electrode matrix.
AuNPs enhance this conductive network through multiple mechanisms: (1) they act as electron transfer bridges between adjacent graphene sheets, minimizing inter-sheet contact resistance; (2) their metallic character provides localized regions of enhanced electric field intensity, facilitating electron tunneling at the biomolecule-electrode interface; and (3) their catalytic activity lowers activation energy barriers for redox reactions involving electroactive species [58] [55].
In biosensing applications, this coordinated charge transport system significantly improves signal-to-noise ratios by maximizing faradaic currents while minimizing non-faradaic background signals. When target analytes bind to recognition elements immobilized on the nanocomposite surface, subsequent redox reactions generate amplified current responses due to the optimized electron transfer pathway to the underlying electrode [60]. This amplification mechanism enables sensitive detection of agricultural relevant compounds including pesticides, fungal toxins, and pathogen biomarkers [57] [3].
Nanocomposite Fabrication and Functionalization
Synthesis of Graphene-AuNP Hybrid Materials
The fabrication of high-performance graphene-AuNP nanocomposites requires precise control over material synthesis and assembly to maximize synergistic effects. Multiple well-established protocols exist for creating these hybrid materials, each offering distinct advantages for specific biosensing applications.
Graphene Oxide (GO) and Reduced Graphene Oxide (rGO) Synthesis typically begins with graphite oxidation using modified Hummer's method, which introduces oxygen-containing functional groups (hydroxyl, epoxy, carboxyl) that facilitate subsequent processing and functionalization [60]. These groups enable excellent water dispersibility and provide anchoring sites for AuNP attachment. Reduction of GO produces rGO with partially restored electrical conductivity while retaining sufficient functional groups for biomolecule immobilization [56]. Thermal, chemical, or electrochemical reduction methods allow tunable control over the final material's electronic properties.
AuNP Synthesis and Assembly most commonly employs the citrate reduction method, where trisodium citrate acts as both reducing agent and stabilizer to produce monodisperse nanoparticles with controllable sizes (5-50 nm) [58]. For graphene composite formation, three primary assembly strategies are employed:
- In-situ growth: AuNP precursors (HAuCl₄) are reduced directly on GO/rGO surfaces, leveraging the material's inherent reducing capability or using additional reducing agents. This approach typically yields strong bonding and uniform nanoparticle distribution [58].
- Ex-situ deposition: Pre-synthesized, functionalized AuNPs are attached to graphene surfaces through covalent bonding, electrostatic interactions, or linker molecules (e.g., cysteamine, dithiocarbamates) [60].
- Self-assembly: Thiol-functionalized graphene sheets spontaneously organize with AuNPs through strong Au-S covalent bonds, creating highly ordered nanostructures with precise interparticle spacing [58].
Table 2: Comparison of Graphene-AuNP Nanocomposite Fabrication Methods
| Fabrication Method | Procedure | Advantages | Limitations | Best Applications |
|---|---|---|---|---|
| In-situ Growth | Chemical reduction of HAuCl₄ on GO/rGO surface | Strong bonding; Uniform distribution; Simple procedure | Limited control over AuNP size distribution | General purpose biosensors; High-volume production |
| Ex-situ Deposition | Attachment of pre-formed AuNPs to graphene | Precise AuNP size control; Customizable surface chemistry | Potential aggregation; Complex multi-step process | Specialized sensors requiring specific AuNP properties |
| Electrochemical Deposition | Electrochemical reduction of Au precursors on electrode | Direct electrode modification; Controlled film thickness | Requires specialized equipment; Limited scalability | Miniaturized sensors; Lab-on-chip devices |
| Self-Assembly | Spontaneous organization via chemical affinity | Highly ordered structures; Molecular-level precision | Sensitive to reaction conditions; Reproducibility challenges | High-precision sensing; Fundamental studies |
The choice of fabrication method significantly influences nanocomposite morphology, stability, and electrochemical performance. For example, in-situ growth typically produces smaller AuNPs (5-15 nm) with dense surface coverage, while ex-situ deposition allows precise control over nanoparticle size and shape at the expense of potentially lower attachment density [60]. Material characterization through techniques such as transmission electron microscopy (TEM), X-ray photoelectron spectroscopy (XPS), Raman spectroscopy, and electrochemical impedance spectroscopy (EIS) is essential for verifying successful nanocomposite formation and optimizing synthesis parameters [58].
Electrode Modification and Biomolecule Immobilization
The successful implementation of graphene-AuNP nanocomposites in electrochemical biosensors requires controlled electrode modification and efficient biomolecule immobilization strategies that preserve biological recognition capabilities while ensuring electrical connectivity.
Electrode Modification Protocols typically employ drop-casting, electrophoretic deposition, or electrochemical reduction techniques to apply nanocomposite materials to electrode surfaces [60]. Screen-printed carbon electrodes (SPCEs) have gained popularity for agricultural biosensing due to their low cost, disposability, and suitability for field analysis [60]. A standard modification procedure involves:
- Electrode pretreatment: Electrochemical cleaning (e.g., cyclic voltammetry in H₂SO₄) or plasma treatment to activate surface functional groups.
- Nanocomposite deposition: Application of optimized graphene-AuNP dispersion (typically 2-5 μL) followed by drying under controlled conditions.
- Surface stabilization: Optional cross-linking or polymer encapsulation to enhance nanocomposite adhesion and prevent leaching during operation.
For example, researchers developing a paraquat sensor demonstrated that sequential modification—first with GO-AuNPs via drop-casting, followed by electrophoretic deposition of poly(3-aminobenzoic acid)—created a stable, hierarchical structure with exceptional analytical performance [60].
Biomolecule Immobilization Strategies leverage the diverse binding capabilities of graphene-AuNP nanocomposites to attach specific recognition elements including enzymes, antibodies, aptamers, and DNA probes [57] [58]. The selection of immobilization chemistry depends on the biological recognition element and the required orientation, density, and stability:
- Enzyme immobilization: Physical adsorption onto graphene surfaces, covalent attachment via carbodiimide chemistry, or entrapment within polymer matrices coexisting with nanocomposites [57]. Acetylcholinesterase (AChE) has been extensively used for organophosphate pesticide detection, where enzyme inhibition by pesticides generates measurable signal changes [57].
- Antibody attachment: Oriented immobilization through Protein A-modified AuNPs, covalent bonding to carboxylic acid groups on graphene, or non-specific adsorption [59]. Proper orientation is critical for maintaining antigen-binding capacity.
- Aptamer functionalization: Thiol-modified aptamers form stable Au-S bonds with AuNPs, while π-π stacking interactions with graphene basal planes provide additional anchoring [58]. This dual-binding approach enhances aptamer stability and surface coverage.
The immobilization process must preserve biological activity while ensuring proximity to the electrode surface for efficient electron transfer. Optimization typically involves controlling surface density, implementing blocking agents to minimize non-specific binding, and verifying biological functionality after immobilization [58] [59].
Experimental Protocols for Agricultural Biosensing
Detection of Pesticide Residues
Organophosphate and other pesticide residues represent significant concerns in agricultural safety, requiring sensitive detection methods for environmental monitoring and food safety assurance. Graphene-AuNP based biosensors have demonstrated exceptional capability for detecting compounds such as chlorpyrifos (CPS) and paraquat at concentrations far below regulatory limits [57] [60].
Protocol 1: Acetylcholinesterase (AChE)-Based Chlorpyrifos Detection
This enzyme-based biosensor leverages the inhibition of AChE activity by organophosphate pesticides for highly sensitive detection [57].
Sensor Fabrication:
- Prepare graphene-AuNP nanocomposite through in-situ growth method (Section 3.1).
- Modify SPCE with 3 μL nanocomposite dispersion and dry at room temperature.
- Immobilize AChE enzyme (0.5 U/μL) onto modified electrode via drop-casting.
- Block non-specific sites with 1% BSA for 30 minutes.
- Wash with phosphate buffer (pH 7.4) to remove unbound components.
Measurement Procedure:
- Incubate modified electrode with sample solution containing CPS for 10-15 minutes.
- Transfer to electrochemical cell containing acetylthiocholine substrate (1 mM) in phosphate buffer.
- Record electrochemical response using differential pulse voltammetry (DPV) or square wave voltammetry (SWV).
- Quantify CPS concentration based on inhibition percentage relative to control measurements.
Performance Characteristics: This approach typically achieves detection limits of 0.1-1 μg/L with linear range from 1-100 μg/L, adequately sensitive for monitoring maximum residue limits in food crops [57]. The sensor shows high specificity for organophosphates over other pesticide classes due to enzyme mechanism.
Protocol 2: Direct Electrochemical Detection of Paraquat
This sensor exploits the inherent redox activity of paraquat, with graphene-AuNP nanocomposites enhancing the electron transfer rate and adsorption capacity [60].
Sensor Fabrication:
- Synthesize GO-AuNP composite and modify SPCE as described in Section 3.2.
- Electropolymerize 3-aminobenzoic acid (10 mM in H₂SO₄) onto GO-AuNP/SPCE via cyclic voltammetry (15 cycles from -0.2 to 2.0 V at 50 mV/s).
- Characterize modified electrode using SEM and electrochemical impedance spectroscopy.
Measurement Procedure:
- Prepare paraquat standards or sample extracts in Britton-Robinson buffer (pH 4.0).
- Accumulate paraquat on electrode surface at open circuit for 120 seconds with stirring.
- Record DPV measurements from -0.8 to -0.2 V with pulse amplitude 50 mV, pulse width 50 ms.
- Quantify paraquat concentration from reduction peak current at approximately -0.65 V.
Performance Characteristics: This method demonstrates exceptional sensitivity with LOD of 0.116 μg/L and linear range from 0.001-100 μM, successfully applied to natural water samples with minimal interference [60].
Detection of Plant Pathogens and Mycotoxins
Early detection of plant pathogens and their associated mycotoxins is crucial for preventing agricultural losses and ensuring food safety. Graphene-AuNP biosensors offer rapid, sensitive alternatives to conventional laboratory methods such as ELISA and PCR [3].
Protocol 3: Aptamer-Based Detection of Fungal Toxins
This protocol describes zearalenone (ZEN) detection using an aptamer-functionalized graphene-AuNP platform, adaptable to other mycotoxins with appropriate recognition elements [61].
Sensor Fabrication:
- Prepare RGO-AuNP nanocomposite through chemical reduction method.
- Modify gold electrode with nanocomposite layer via drop-casting.
- Immobilize thiol-modified ZEN aptamer (5 μM) onto surface through self-assembly (12-16 hours at 4°C).
- Block with 6-mercapto-1-hexanol (1 mM) for 2 hours to passivate uncovered Au surfaces.
- Characterize stepwise modification using EIS and CV.
Measurement Procedure:
- Incubate aptasensor with ZEN standards or sample extracts for 30 minutes.
- Measure electrochemical response using DPV in [Fe(CN)₆]³⁻/⁴⁻ solution.
- Correlate signal reduction with ZEN concentration due to aptamer conformation change.
- Regenerate sensor with mild alkaline solution (pH 9.0) for repeated measurements.
Performance Characteristics: Aptasensors typically achieve LOD values of 0.1-1 μg/kg with wide linear ranges, suitable for monitoring mycotoxin levels in grains and oilseeds [61] [3].
Protocol 4: Immunosensor for Plant Pathogen Detection
This protocol outlines antibody-based detection for fungal pathogens like Sclerotinia sclerotiorum, a major threat to oilseed crops [3].
Sensor Fabrication:
- Modify SPCE with graphene-AuNP nanocomposite to enhance surface area and conductivity.
- Immobilize capture antibodies against pathogen-specific antigens via EDC/NHS chemistry.
- Block with 1% casein or BSA for 60 minutes.
- Apply secondary antibodies conjugated with horseradish peroxidase (HRP) for enzymatic amplification.
Measurement Procedure:
- Incubate sensor with sample extract (30 minutes) followed by washing.
- Add HRP substrate solution (H₂O₂ with hydroquinone mediator).
- Measure amperometric response at -0.2 V vs. Ag/AgCl.
- Quantify pathogen concentration from calibration curve.
Performance Characteristics: These immunosensors can detect pathogen biomarkers at concentrations as low as 1-10 ng/mL, enabling early diagnosis before visual symptoms appear [3].
Performance Analysis and Applications
Analytical Performance Comparison
The integration of graphene-AuNP nanocomposites into electrochemical biosensors has yielded exceptional analytical performance across diverse agricultural applications. The following table summarizes representative performance metrics for different sensor configurations targeting various agricultural analytes.
Table 3: Performance Comparison of Graphene-AuNP Based Biosensors for Agricultural Monitoring
| Target Analyte | Recognition Element | Detection Method | Linear Range | Limit of Detection | Application Example |
|---|---|---|---|---|---|
| Chlorpyrifos | Acetylcholinesterase enzyme | DPV | 0.01-100 μg/L | 0.005 μg/L | Food samples, water sources [57] |
| Paraquat | Direct detection (redox active) | DPV | 0.001-100 μM | 0.45 nM (0.116 μg/L) | Natural water, tap water [60] |
| PCB77 | DNA aptamer | DPV | 1 pg/L - 10 μg/L | 0.1 pg/L | Environmental samples [58] |
| Zearalenone | Aptamer | EIS | 0.1-100 μg/kg | 0.05 μg/kg | Grain samples [61] |
| Plant Pathogens | Antibody | Amperometry | 1-1000 ng/mL | 0.5 ng/mL | Oilseed crop leaves [3] |
The exceptional sensitivity demonstrated across these applications highlights the effective signal amplification achieved through graphene-AuNP nanocomposites. Detection limits frequently surpass regulatory requirements, enabling early warning systems for agricultural contaminants. For example, the LOD for chlorpyrifos detection (0.005 μg/L) is significantly lower than the maximum residue limits established by international food safety authorities [57]. Similarly, the ultra-sensitive detection of PCB77 (0.1 pg/L) satisfies the strict exposure thresholds set by environmental protection agencies [58].
The Researcher's Toolkit: Essential Materials and Reagents
Successful implementation of graphene-AuNP enhanced biosensors requires specific materials and reagents optimized for agricultural applications. The following table outlines essential components and their functions in sensor development and deployment.
Table 4: Essential Research Reagents for Graphene-AuNP Biosensor Development
| Category | Specific Materials | Function | Application Notes |
|---|---|---|---|
| Nanomaterials | Graphite powder, Graphene oxide, Chloroauric acid (HAuCl₄) | Sensor platform construction | Purity and source significantly impact reproducibility [60] [56] |
| Recognition Elements | Acetylcholinesterase, Specific aptamers, Antibodies | Target capture and specificity | Require proper immobilization to maintain activity [57] [58] |
| Electrochemical Substrates | Potassium ferricyanide, Acetylthiocholine, H₂O₂ with mediators | Signal generation | Selection depends on transduction mechanism [57] [60] |
| Buffer Systems | Phosphate buffer (pH 7.4), Britton-Robinson buffer, Acetate buffer | Optimal biorecognition environment | pH and ionic strength critically affect performance [60] [3] |
| Linker Molecules | Cysteamine, APTES, EDC/NHS chemistry | Biomolecule immobilization | Control orientation and density of recognition elements [58] [59] |
Future Perspectives and Research Challenges
Despite significant advances in graphene-AuNP based biosensing platforms, several challenges remain for widespread agricultural implementation. Current limitations include signal interference from complex plant matrices, finite sensor stability under field conditions, and requirements for sophisticated data interpretation [3]. Future research directions should focus on several key areas:
Multiplexed Detection Platforms integrating multiple recognition elements on a single electrode array would enable simultaneous monitoring of different pathogens, pesticides, or toxins, providing comprehensive crop health assessment [54]. The versatile surface chemistry of graphene-AuNP nanocomposites makes them ideal foundations for such multidimensional sensing platforms.
Advanced Signal Amplification Strategies incorporating additional nanomaterial layers or catalytic cycles could further enhance detection sensitivity. Emerging approaches include DNA nanotechnology for programmed assembly, hybridization chain reactions for exponential signal amplification, and integration with microfluidic systems for automated sample processing [55] [59].
Field-Deployable Device Integration represents the ultimate translation goal for agricultural biosensors. Recent innovations in miniaturized potentiostats, wireless data transmission, and power management systems are gradually overcoming barriers to field deployment [54] [3]. The compatibility of graphene-AuNP sensors with screen-printed electrode technology facilitates disposable, cost-effective form factors suitable for widespread monitoring.
The integration of artificial intelligence for data interpretation and IoT connectivity for real-time monitoring will further enhance the practical utility of these biosensing systems, potentially revolutionizing agricultural management practices through precise, data-driven decision support [3]. As these technologies mature, graphene-AuNP enhanced biosensors are poised to become indispensable tools for sustainable agriculture, enabling proactive disease management, optimized pesticide usage, and enhanced food safety assurance.
Strategies for Improving Sensor Stability, Biocompatibility, and Lifespan
The integration of electrochemical biosensors into agricultural research represents a paradigm shift towards data-driven, precision farming. These sensors enable the real-time monitoring of a wide range of analytes, from soil nutrients and environmental pollutants to plant pathogens and foodborne contaminants [16]. However, the transition from laboratory prototypes to reliable field-deployable tools is hindered by three interconnected challenges: stability under variable environmental conditions, biocompatibility to minimize adverse biological responses, and sufficient lifespan for practical long-term monitoring [62] [16]. This technical guide synthesizes current advances and provides detailed methodologies to address these critical limitations, providing a roadmap for researchers developing robust sensing systems for agricultural applications.
Core Challenges in Agricultural Biosensing
The agricultural environment presents a uniquely demanding set of conditions for biosensors. Key challenges include:
- Biofouling: The nonspecific adsorption of proteins, cells, bacteria, and other organic molecules onto sensor surfaces can rapidly degrade signal accuracy by obstructing the interaction between the target analyte and the biorecognition element [63]. This is a significant issue in soil, plant sap, and food matrices.
- Foreign Body Response (FBR): For implantable sensors in plants or animals, the natural immune response to a foreign material can lead to the formation of fibrotic tissue, isolating the sensor and restricting analyte access [62] [63].
- Material Degradation: The mechanical integrity and electrochemical performance of sensor materials can be compromised by prolonged exposure to moisture, UV radiation, enzymatic activity, and fluctuating pH levels common in agricultural settings [16].
- Signal Drift and Calibration Loss: Environmental stressors and bioreceptor instability can lead to signal drift, making continuous, long-term measurements unreliable without frequent, impractical recalibration [62].
Overcoming these barriers requires a multi-faceted strategy focusing on advanced materials, smart engineering, and rigorous testing protocols.
Material Design and Interface Engineering
The interface between the sensor and its biological environment is the primary determinant of its performance. Engineering this interface is critical for enhancing stability and biocompatibility.
Advanced Nanocomposites and Conductive Polymers
The strategic use of nanocomposites and polymers can significantly improve electron transfer, bioreceptor immobilization, and structural stability.
- Nanomaterial-Enhanced Electrodes: Nanomaterials such as graphene, carbon nanotubes, and metal nanoparticles are extensively used to create high-surface-area, three-dimensional (3D) structures for probe immobilization. These structures enhance sensitivity and provide a stable scaffold for bioreceptors. For instance, a multi-dimensional hollow
C@SnO2nanocomposite has been demonstrated as an excellent electrode substrate, offering a large specific surface area, excellent electron transport properties, and robust structural stability that inhibits volume expansion during electrochemical reactions [64]. - Conductive Hydrogels and Polymers: Materials like poly(3,4-ethylenedioxythiophene) (PEDOT) and polyaniline offer high conductivity and biocompatibility. Hydrogels, in particular, create a hydrating, bio-friendly microenvironment that protects both the sensor surface and the surrounding tissue [65]. They can be designed as
hydrogel nano(bio)composite sensorsthat allow for real-time monitoring and detection of multiple parameters simultaneously [65].
Smart Coatings for Biocompatibility and Anti-Biofouling
Creating a physical and chemical barrier at the sensor interface is a primary defense mechanism.
- Albumin-Graphene Coating: A breakthrough coating technology involves a cross-linked lattice of Bovine Serum Albumin (BSA) and functionalized graphene. The BSA forms a natural barrier that blocks the non-specific binding of biofluids and contaminants, while the functionalized graphene ensures efficient electrical signaling. This coating has been shown to resist biofilm formation by bacteria like P. aeruginosa, prevent fibroblast adhesion, and suppress pro-inflammatory immune responses, maintaining sensor functionality in complex human plasma for over three weeks [63].
- Zwitterionic Polymer Coatings: Polymers with zwitterionic (mixed positive and negative charges) groups create a super-hydrophilic surface that strongly binds water molecules, forming a physical and energetic barrier to protein adsorption and cell attachment. As demonstrated by Lv et al. (2024), coatings such as epoxy propyl dimethyl ammonium chloride can significantly improve sensor stability for extended continuous use in biological fluids like interstitial fluid [66].
- Biodegradable Materials: For single-use applications, biodegradable materials can eliminate the need for sensor removal surgery, reducing environmental impact and operational complexity [62].
Table 1: Summary of Key Coating Strategies and Their Performance
| Coating Strategy | Key Components | Mechanism of Action | Reported Performance |
|---|---|---|---|
| Albumin-Graphene Matrix [63] | BSA, Functionalized Graphene | Creates a natural barrier against non-specific binding; suppresses immune response. | >3 weeks stability in plasma; resists biofouling and fibroblast adhesion. |
| Zwitterionic Polymer [66] | Epoxy propyl dimethyl ammonium chloride | Forms a hydration layer via electrostatic interactions; reduces protein adsorption. | Improved stability for long-term continuous use in interstitial fluid. |
| Hydrogel-Based [65] | Polymeric networks (e.g., PEDOT, PVA) | Provides a hydrating, biocompatible microenvironment; reduces foreign body response. | Enables real-time monitoring; suitable for implantable devices. |
Experimental Protocols for Validation
Rigorous and standardized experimental validation is essential for quantifying the improvements offered by new materials and designs.
Protocol for Assessing Anti-Biofouling Performance
Objective: To evaluate the efficacy of a sensor coating in resisting nonspecific protein adsorption and bacterial biofilm formation.
Materials:
- Coated and uncoated (control) sensor electrodes.
- Complex protein solution (e.g., 10% fetal bovine serum in PBS or plant sap extract).
- Bacterial culture (e.g., Pseudomonas aeruginosa in LB broth).
- Electrochemical impedance spectroscopy (EIS) setup.
Methodology:
- Baseline Measurement: Record the EIS spectrum (e.g., 0.1 Hz to 100 kHz) for all sensors in a clean PBS buffer.
- Protein Exposure: Incubate sensors in the protein solution for a predetermined period (e.g., 1-24 hours) at 37°C with gentle agitation.
- Post-Protein EIS: Gently rinse the sensors with PBS and record the EIS spectrum again. An increase in charge-transfer resistance (
R_ct) indicates protein adsorption. - Biofilm Challenge: Incubate sensors in the bacterial culture for 24-48 hours.
- Analysis: Use microscopy (e.g., SEM, confocal) to visualize biofilm formation and EIS to quantify the performance degradation. The percentage change in
R_ctfor coated vs. uncoated sensors quantifies the coating's effectiveness [63] [66].
Protocol for In Vivo Biocompatibility and Lifespan Testing
Objective: To determine the sensor's functional lifespan and tissue response in a live organism (e.g., a plant or animal model).
Materials:
- Implantable biosensors with the coating/design of interest.
- Suitable animal model (e.g., rodent) or plant model (e.g., tomato, oilseed rape).
- Histology equipment.
Methodology:
- Sensor Implantation: Surgically implant the sensor into the target tissue (e.g., subcutaneous space, stem vascular tissue).
- Continuous Monitoring: Record the sensor's signal (e.g., amperometric current for a specific analyte) at regular intervals over several weeks.
- Calibration Check: Periodically measure the sensor's response to a standard solution in situ or post-explantation to check for signal drift.
- Endpoint Analysis: After a set period (e.g., 3+ weeks), euthanize the organism and explant the sensor along with the surrounding tissue.
- Histological Analysis: Section and stain the tissue (e.g., with H&E and Masson's trichrome) to evaluate the immune cell presence (inflammation) and collagen deposition (fibrous capsule thickness). A thinner capsule and fewer inflammatory cells indicate superior biocompatibility [62] [63].
Power Management and Data Integrity
For long-term, in-field deployment, ensuring a stable power supply and reliable data is paramount.
- Energy Harvesting and Self-Powered Systems: Despite advances in energy harvesting, power management remains a critical challenge for implantable and wireless sensors [62]. A promising solution is the development of self-powered sensors based on technologies like enzyme biofuel cells (EBFCs). These systems use redox reactions to convert chemical energy from biofuels (e.g., glucose in plant sap) into electrical energy, powering the sensor for target detection without an external power source [64].
- Signal Amplification and AI-Assisted Data Interpretation: To combat signal drift and matrix interference, integrating signal amplification strategies directly into the sensor design is highly effective. The Catalytic Hairpin Assembly (CHA) is a non-enzymatic, isothermal nucleic acid amplification method that can significantly enhance detection sensitivity [64]. Furthermore, coupling sensors with mobile platforms and AI/ML analytics can help correct for baseline drift, distinguish target signals from noise, and provide actionable insights for decision support systems [16].
Table 2: The Scientist's Toolkit: Essential Research Reagents and Materials
| Item / Reagent | Function in R&D | Application Example |
|---|---|---|
| Bovine Serum Albumin (BSA) | Blocking agent; matrix for cross-linked anti-fouling coatings. | Primary component in albumin-graphene anti-fouling coatings [63]. |
| Functionalized Graphene Oxide | Provides high conductivity and a platform for biomolecule immobilization. | Enhances electron transfer in 3D electrode structures and composite coatings [63]. |
| Poly(3,4-ethylenedioxythiophene) (PEDOT) | Conductive polymer for electrode coating and hydrogel formation. | Improves charge transfer at the working electrode; used in microneedle sensors [66] [65]. |
| Zwitterionic Polymers | Creates ultra-low fouling surfaces via strong hydration. | Coating for implantable sensors to resist protein and cell adhesion [66]. |
| Catalytic Hairpin Assembly (CHA) Probes | Provides isothermal, enzymatic signal amplification for high sensitivity. | Detecting ultra-low concentrations of specific miRNA or DNA targets [64]. |
| Prussian Blue | Electron mediator for enhancing H₂O₂-based detection. |
Used in amperometric biosensors (e.g., with glucose oxidase) to lower operating potential [66]. |
| SiO₂ Template Nanoparticles | Sacrificial template for creating hollow nanostructures. | Synthesis of multi-dimensional hollow C@SnO₂ nanocomposites for self-powered sensors [64]. |
Visualization of Signaling and Response Pathways
Understanding the biological signaling pathways involved in the foreign body response is crucial for designing effective countermeasures.
The path to widespread adoption of electrochemical biosensors in agriculture hinges on overcoming the intertwined challenges of stability, biocompatibility, and lifespan. As detailed in this guide, the strategic use of smart coatings like albumin-graphene matrices, the development of 3D nanostructured electrodes, and the integration of self-powered systems and AI-driven data analysis represent the forefront of this endeavor [62] [16] [63]. Future research must focus on the creation of standardized testing protocols, the exploration of fully biodegradable sensor platforms for single-use applications, and the seamless integration of these devices into agricultural Internet of Things networks. By addressing these core engineering challenges, electrochemical biosensors will fully realize their potential as powerful tools for sustainable precision agriculture, enhancing food safety, optimizing resource use, and strengthening global food security.
Electrochemical biosensors have emerged as promising tools for advancing agricultural research, offering the potential for rapid, on-site detection of pathogens, toxins, and other critical analytes directly in field conditions. These sensors merge biological recognition elements with electrochemical transducers, converting specific biochemical interactions into quantifiable electrical signals such as current, potential, or impedance [67]. Their relevance to agricultural applications is significant, enabling early detection of devastating crop diseases like downy mildew in oilseed rape or soybean rust before visible symptoms manifest, thereby preventing substantial yield losses that can reach 30-50% [15] [3]. Despite their transformative potential, the widespread deployment of these sophisticated analytical tools in agricultural settings faces three fundamental engineering challenges: device miniaturization for field-portable operation, managing power requirements for sustained monitoring, and achieving cost-effective production for scalable implementation. This technical guide examines these core hurdles and outlines current innovative approaches aimed at overcoming these barriers to practical adoption.
Miniaturization: Balancing Performance with Portability
The drive toward miniaturizing electrochemical biosensors is motivated by the need for portable, field-deployable devices that can provide real-time monitoring in agricultural environments. Miniaturization efforts focus primarily on developing compact sensing architectures and integrating microfluidic systems for sample handling.
Microfluidic Integration and Sensor Architectures
Lab-on-a-Chip (LoC) systems represent the forefront of miniaturization, integrating multiple laboratory functions—including sample preparation, mixing, separation, and detection—onto a single microfluidic chip that handles extremely small fluid volumes [25]. These systems are particularly valuable for agricultural applications where sample complexity and the need for rapid, on-site analysis present significant challenges. When combined with electrochemical detection, LoC platforms enable sensitive, portable, and cost-effective analysis of complex agricultural matrices like soil extracts or plant sap with accuracy and repeatability [25]. The core advantage of these integrated systems lies in their ability to efficiently manage typical field challenges such as matrix interferences and low analyte concentrations by combining sample preparation, separation, and detection into a single, compact architecture [25].
Recent innovations in electrode fabrication have further advanced miniaturization capabilities. Laser ablation techniques now enable the creation of electrodes with customizable geometries and microlevel resolutions [68]. This approach, when combined with unconventional materials like laminated gold leaves on polyvinyl chloride (PVC) adhesive sheets, allows for rapid production of highly conductive electrodes with large surface areas suitable for biomolecule immobilization [68]. Similarly, laser-scribing methods can directly pattern conductive graphene circuits on flexible substrates, creating miniaturized sensors without the need for complex lithography [69]. These fabrication advances are particularly relevant for agricultural applications where flexible, durable sensor platforms are needed to monitor crops in field conditions.
Technical Challenges in Miniaturized Systems
Despite these advances, miniaturization introduces significant technical constraints that impact sensor performance:
- Complex Fabrication Requirements: Techniques like photolithography and chemical vapor deposition (CVD), while producing highly precise and reproducible electrodes, require expensive equipment, cleanroom facilities, and specific chemicals, increasing complexity and cost [68].
- Signal Interference: Miniaturized sensors are more susceptible to signal interference from complex plant matrices, which can reduce detection accuracy in real-world agricultural samples [15] [3].
- Limited Functional Integration: Incorporating multiple sensing components, sample pretreatment steps, and reference electrodes within a small footprint remains challenging, often requiring trade-offs between functionality and size [67] [70].
Table 1: Miniaturization Approaches and Their Limitations in Agricultural Biosensors
| Approach | Key Features | Agricultural Applications | Limitations |
|---|---|---|---|
| Lab-on-a-Chip (LoC) | Integrates sample preparation, separation, detection; minimal reagent use | On-site pathogen detection (e.g., Phakopsora pachyrhizi in soybeans) | Complex fabrication; matrix interference in plant samples |
| Laser-Ablated Electrodes | Customizable geometries; microlevel resolution; rapid prototyping | Field-deployable sensors for oilseed crop diseases | Limited conductivity with some materials (e.g., gold leaf) |
| Screen-Printed Electrodes | Mass production capability; various substrates (ceramics, polymers) | Disposable pesticide sensors; soil contaminant monitoring | Reproducibility challenges; ink impurities affect performance |
| Wearable Sensors | Direct plant integration; continuous monitoring | Crop health monitoring; stress biomarker detection | Power constraints; environmental vulnerability |
Power Management: Enabling Field-Deployable Operation
Power requirements present a critical constraint for electrochemical biosensors intended for remote or extended agricultural monitoring. Efficient power management strategies focus on both reducing sensor energy demands and exploring alternative power sources suitable for field deployment.
Low-Power Electrochemical Sensing Modalities
Different electrochemical sensing techniques exhibit varying power requirements, with selection depending on the specific agricultural application and operational context:
- Amperometric Sensors: These sensors maintain a constant potential while measuring current, offering relatively low power consumption that makes them suitable for continuous monitoring of metabolites like glucose or lactate in plant tissues [67]. Their simplicity and efficiency have established them as the foundation for most portable agricultural sensors, including commercially successful platforms like personal glucose meters which demonstrate the potential for long-term operation with minimal power resources [67].
- Voltammetric Techniques: Methods such as cyclic voltammetry and differential pulse voltammetry offer enhanced sensitivity for detecting proteins, nucleic acids, and pathogens but typically require more sophisticated instrumentation and greater power resources due to their potential scanning protocols [67]. While these techniques provide valuable data for detecting agricultural pathogens like Sclerotinia sclerotiorum at ultra-low concentrations, their power requirements often limit their suitability for truly long-term field deployment [15].
- Impedimetric Sensors: Electrochemical impedance spectroscopy (EIS) measures non-faradaic resistance and capacitance properties, requiring minimal power as it uses small amplitude AC voltages [70]. This inherent efficiency gives impedance biosensors significant potential for development into portable environmental monitoring systems, though challenges with non-specific binding in complex plant samples remain [70].
Power Source Innovations
For agricultural applications requiring extended deployment, innovative power solutions are emerging:
- Integration with Energy Harvesting Systems: Some research platforms now incorporate solar cells or bioelectrochemical systems that generate power from soil microbial activity, creating self-sustaining sensor networks for field conditions [70].
- Smartphone-Based Power Management: The integration of electrochemical sensors with smartphones leverages the phone's battery while utilizing its computational capabilities, creating an efficient all-in-one solution that simplifies power management for field researchers [25].
- Low-Power Wireless Communication: For sensors integrated into Agricultural Internet of Things (IoT) networks, power-efficient communication protocols like LoRaWAN or NB-IoT enable extended operation with minimal energy draw, facilitating real-time crop health monitoring across large agricultural areas [15] [3].
Table 2: Power Requirements and Management Strategies for Agricultural Biosensors
| Sensor Type | Power Consumption | Optimal Use Cases | Power Management Strategies |
|---|---|---|---|
| Amperometric | Low (constant potential) | Continuous metabolite monitoring; pesticide detection | Battery-powered with sleep modes; ideal for portable devices |
| Impedimetric | Low (small AC voltage) | Pathogen detection; soil quality assessment | Energy harvesting from environment; low-power circuitry |
| Voltammetric | Moderate to high (potential scanning) | Research-grade analysis; multiplexed detection | Rechargeable batteries; smartphone integration |
| Potentiometric | Low (zero current) | Soil pH monitoring; ion detection (e.g., K+, NO3-) | Long-term battery operation; passive sensing approaches |
Cost-Effective Production: Scaling Agricultural Biosensors
Achieving cost-effective manufacturing is essential for widespread adoption of electrochemical biosensors in agriculture, where profit margins are often narrow and monitoring needs extensive. Recent advances in fabrication technologies and materials science have enabled significant progress toward this goal.
Innovative Fabrication Approaches
Several manufacturing techniques have emerged as particularly promising for producing low-cost, disposable electrochemical sensors suitable for agricultural applications:
- Laser Ablation and Gold Leaf Lamination: This innovative approach combines 24-karat gold leaves with low-cost polyvinyl chloride (PVC) adhesive sheets, patterned using laser ablation to create electrodes with customizable geometries at microlevel resolutions [68]. The method significantly reduces material costs compared to traditional gold deposition techniques like sputtering or evaporation while maintaining excellent electrical conductivity and biocompatibility for immobilizing biorecognition elements like antibodies or aptamers [68].
- Screen and Stencil Printing: These techniques enable mass production of planar electrochemical sensors using conductive inks or pastes on various substrates including ceramics, polymers, and paper [68] [69]. While screen printing offers scalability and cost-effectiveness for producing disposable sensors, challenges with reproducibility due to screen imperfections and potential interference from organic binders in the inks remain limitations [68]. Stencil printing provides a simpler alternative using cut patterns to define electrode structures, though with potentially lower resolution [69].
- Pencil Drawing and Laser-Scribing: For extreme cost reduction, pencil drawing with graphite cores on paper substrates creates simple conductive traces without requiring any specialized equipment [69]. Similarly, laser-scribing can convert polymer surfaces into conductive graphene patterns, enabling direct fabrication of electrodes on flexible materials [69]. These approaches offer particular promise for resource-limited agricultural settings where sophisticated manufacturing capabilities are unavailable.
- Additive Manufacturing (3D Printing): This technology provides precise control over electrode geometry, enabling creation of complex microstructures and flexible sensors while integrating multiple sensing components to reduce manufacturing costs [68]. Although challenges remain regarding resolution and material conductivity, 3D printing holds significant promise for cost-effective, scalable production of disposable biosensors for various agricultural applications [68].
Material Selection and Nanomaterial Integration
The choice of electrode materials significantly impacts both sensor performance and production costs:
- Gold Nanomaterials: While gold offers excellent electrical conductivity, chemical stability, and biocompatibility, traditional fabrication techniques like chemical vapor deposition (CVD) are costly and environmentally challenging [68]. The development of alternative fabrication methods using gold leaves or gold nanoparticles (AuNPs) integrated with cost-effective substrates has dramatically reduced material expenses while maintaining performance benefits [68] [25].
- Carbon-Based Materials: Graphene, graphene oxide (GO), and carbon nanotubes provide high surface-to-volume ratios and excellent electrical properties at lower cost than precious metals [25]. These materials can be incorporated into conductive inks for printing technologies or used as electrode modifiers to enhance sensitivity through increased surface area and improved electron transfer kinetics [69] [25].
- Paper and Flexible Substrates: Paper-based electrochemical sensors offer an extremely low-cost platform ideal for disposable applications in agricultural monitoring [69]. The inherent porosity of paper enables capillary-driven fluid transport without external pumping, simplifying device design and reducing costs further [69].
Low-Cost Biosensor Fabrication Ecosystem
Integrated Experimental Framework: A Case Study in Agricultural Monitoring
To illustrate how these engineering challenges are addressed in practice, this section presents a detailed experimental framework for developing a biosensor targeting agricultural pathogens, incorporating specific protocols for fabrication, testing, and validation.
Fabrication Protocol: Gold Leaf Electrode Production
The following methodology outlines the creation of low-cost, high-performance electrodes suitable for detecting plant pathogens such as Sclerotinia sclerotiorum or Phakopsora pachyrhizi:
Materials Preparation:
- Substrate: Polyvinyl chloride (PVC) adhesive sheets (125 μm thickness)
- Conductive material: 24-karat gold leaves (80 mm × 80 mm)
- Surface treatment: PTFE (polytetrafluoroethylene) dry lubricant spray
- Laser system: CO₂ or fiber laser ablation system
Step-by-Step Fabrication:
- Surface Treatment: Apply PTFE spray to a clean, flat working surface to prevent gold leaf adhesion during processing [68].
- Gold Leaf Lamination: Carefully place the gold leaf onto the PVC adhesive sheet, ensuring smooth, wrinkle-free attachment. Apply even pressure to establish firm contact between layers [68].
- Laser Patterning: Program the laser ablation system with the desired electrode geometry (typically three-electrode configuration: working, reference, and counter electrodes). Execute laser patterning to remove excess gold leaf, creating precisely defined conductive pathways [68].
- Quality Assessment: Verify electrode integrity using scanning electron microscopy (SEM) for surface morphology and 3D profiling for dimensional accuracy [68].
- Electrochemical Activation: Perform cyclic voltammetry in 0.5 M H₂SO₄ solution (potential range: -0.2 to +1.5 V vs. Ag/AgCl) until stable voltammograms appear, indicating proper electrode activation [68].
Performance Validation:
- Conduct electrochemical characterization using 10 mM potassium ferricyanide/ferrocyanide redox couple in PBS (pH 7.4)
- Parameters: Calculate electrode active area using Randles-Sevcik equation; measure electron transfer kinetics through peak separation (ΔEp)
- Acceptance criteria: ΔEp < 80 mV for reversible systems; relative standard deviation (RSD) < 5% for replicate measurements [68]
Sensor Integration and Agricultural Implementation
Following electrode fabrication, the functionalization and integration process enables specific pathogen detection:
Biorecognition Element Immobilization:
- For antibody-based detection: Immobilize pathogen-specific antibodies (e.g., anti-Sclerotinia sclerotiorum) through covalent binding to self-assembled monolayers (SAMs) on gold surfaces using thiol chemistry [15].
- For aptamer-based detection: Modify DNA or RNA aptamers with thiol groups at 5' or 3' ends for oriented immobilization on gold electrodes [15].
- Blocking step: Incubate with 1% BSA for 1 hour to minimize non-specific binding [15].
Magnetic Bead Enhancement (for increased sensitivity):
- Use magnetic beads (e.g., Pathatrix Dual Kit) conjugated with secondary antibodies or aptamers
- Implement magnetic concentration of target pathogens prior to detection to improve limits of detection [68]
- Apply external magnetic field to concentrate bead-target complexes on electrode surface
Signal Measurement:
- Utilize differential pulse voltammetry (DPV) or electrochemical impedance spectroscopy (EIS) for quantitative detection
- DPV parameters: Potential range -0.1 to +0.5 V, pulse amplitude 25 mV, pulse width 50 ms
- EIS parameters: Frequency range 0.1 Hz to 100 kHz, AC amplitude 10 mV [68]
Agricultural Biosensor Development Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful development of electrochemical biosensors for agricultural applications requires carefully selected materials and reagents that address both performance requirements and cost constraints. The following table summarizes key components and their functions in sensor fabrication and operation.
Table 3: Essential Research Reagents and Materials for Agricultural Biosensor Development
| Category | Specific Materials | Function in Biosensor Development | Agricultural Application Examples |
|---|---|---|---|
| Electrode Materials | Gold leaf (24-karat), Carbon inks, Graphene oxide | Create conductive pathways for electron transfer; interface for biorecognition immobilization | Pathogen detection in oilseed crops; soil contaminant monitoring |
| Biorecognition Elements | Antibodies, Aptamers, Molecularly imprinted polymers (MIPs) | Provide specific binding to target analytes (pathogens, toxins) | Detection of Sclerotinia sclerotiorum; aflatoxin monitoring in peanuts |
| Signal Amplification | Magnetic beads, Gold nanoparticles, Enzymes (HRP, GOx) | Enhance detection sensitivity through concentration or catalytic activity | Preconcentration of low-abundance pathogens; signal enhancement for early detection |
| Substrate Materials | PVC sheets, Paper substrates, Flexible polymers | Provide mechanical support for electrode structures; enable disposable formats | Field-deployable sensors; wearable plant health monitors |
| Electrochemical Reagents | Ferri/ferrocyanide redox couple, PBS buffer, H₂SO₄ | Enable electrode characterization and electrochemical measurements | Sensor validation; performance optimization for plant sample matrices |
The development of electrochemical biosensors for agricultural applications continues to advance, with ongoing research addressing the fundamental engineering challenges of miniaturization, power management, and cost-effective production. Emerging trends point toward several promising directions that may further overcome current limitations.
Integration with Digital Agriculture Infrastructure represents a particularly promising pathway. The combination of electrochemical sensors with Internet of Things (IoT) platforms and artificial intelligence (AI)-assisted data interpretation creates opportunities for comprehensive crop health monitoring systems that provide real-time analytics to farmers and researchers [15] [3]. These integrated systems can leverage the low-power capabilities of modern electrochemical sensors while utilizing edge computing for localized data processing, minimizing power-intensive data transmission needs.
Advanced Materials Science continues to drive progress in cost reduction and performance enhancement. The development of biodegradable sensor materials addresses both cost and environmental concerns, while further innovations in nanomaterial integration promise to improve sensitivity without significantly increasing production expenses [15] [3]. Similarly, novel energy harvesting approaches specific to agricultural environments—such as photosynthesis-based power systems or soil microbial fuel cells—may ultimately eliminate battery dependencies for certain applications.
Standardization and Commercialization efforts will be crucial for translating laboratory innovations into practical agricultural tools. The absence of standardized detection protocols currently hinders direct comparison between sensor platforms and validation across different agricultural settings [15]. Addressing this limitation through collaborative initiatives between academic researchers, agricultural experts, and industry partners will accelerate the adoption of electrochemical biosensors in mainstream agricultural practice.
As these technological advances converge, electrochemical biosensors are poised to become increasingly integral to precision agriculture, enabling more sustainable farming practices through targeted intervention, reduced chemical usage, and enhanced crop management capabilities. The ongoing resolution of engineering challenges in miniaturization, power, and cost will ultimately determine the scale and effectiveness of this transformation.
Benchmarking Performance: Validation Against Traditional Methods and Comparative Analysis
Analytical chemistry applied to medical and diagnostic analysis has increasingly focused on developing cost-effective biosensors for monitoring health status or assessing biomarker levels indicative of various diseases [71]. The improvement of technologies for non-invasive biological fluid sampling, signal detection, and computational capabilities has made complete integration of self-standing analytical devices more accessible [71]. In agricultural research, particularly within the context of precision farming and sustainable agriculture, biosensors have emerged as powerful tools for real-time monitoring of plant health, soil quality, and food safety [72] [73].
This technical guide provides an in-depth comparison between two dominant biosensing technologies—electrochemical and optical—focusing on their fundamental principles, performance characteristics, and practical applications in agricultural research. The content is structured to assist researchers, scientists, and development professionals in selecting appropriate biosensing platforms for specific agricultural applications, with particular emphasis on the growing role of electrochemical biosensors in addressing modern farming challenges.
Fundamental Principles and Mechanisms
Electrochemical Biosensors
Electrochemical biosensors operate by measuring changes in electrical parameters—current, voltage, or impedance—that occur when a target analyte interacts with a biological recognition element immobilized on an electrode surface [66] [74]. These sensors typically utilize a three-electrode system consisting of working, reference, and counter electrodes [66]. The biological recognition element, which may be an enzyme, antibody, aptamer, or whole cell, facilitates specific binding with the target analyte, generating an electrochemical signal proportional to the analyte concentration [74].
Key electrochemical detection methods include:
- Amperometry: Measures current changes at a fixed potential, widely used in enzyme-based sensors (e.g., glucose oxidase for glucose detection) [66]
- Potentiometry: Measures potential differences between electrodes when no significant current flows [66]
- Impedance Spectroscopy (EIS): Monitors impedance changes at the electrode interface upon analyte binding [66]
Recent innovations in electrochemical biosensing have focused on nanomaterial integration to enhance sensitivity and selectivity. For instance, gold nanoparticles, graphene, and conductive polymers like PEDOT have been extensively used to modify electrode surfaces, improving charge transfer efficiency and providing larger surface areas for bioreceptor immobilization [66] [3].
Optical Biosensors
Optical biosensors convert biological recognition events into measurable optical signals through various transduction mechanisms including absorbance, fluorescence, chemiluminescence, and refractive index changes [73] [75]. These sensors combine biorecognition molecules with advanced optical transducers to detect a wide array of analytes with high sensitivity and specificity [73].
Major optical biosensing modalities include:
- Surface Plasmon Resonance (SPR): Detects changes in refractive index at a metal-dielectric interface when analytes bind to immobilized recognition elements [71] [73]
- Fluorescence and Chemiluminescence: Employ fluorescent tags or luminescent molecules to generate measurable emission changes in response to analyte binding [71] [73]
- Colorimetric Sensing: Provides visible color changes in the presence of specific analytes, allowing interpretation with minimal equipment [71] [76]
Nanotechnology has significantly advanced optical biosensing capabilities, with quantum dots, gold nanoparticles, and carbon-based nanomaterials enhancing light-matter interactions and enabling ultra-sensitive detection [73]. For agricultural applications, optical biosensors have proven particularly valuable for detecting food contaminants, pathogens, and toxins [73].
Performance Comparison and Technical Specifications
The selection between electrochemical and optical biosensing platforms depends heavily on performance requirements for specific applications. The table below summarizes key technical parameters for both biosensor types:
Table 1: Performance Comparison of Electrochemical and Optical Biosensors
| Parameter | Electrochemical Biosensors | Optical Biosensors |
|---|---|---|
| Sensitivity | Extremely high (e.g., H₂O₂ detection: 14.7 μA/μM; pesticides: 0.029–21 fg/mL) [66] | High (SPR, fluorescence enable trace contaminant detection) [73] |
| Detection Limit | Low LOD (e.g., insulin: 0.01-4 nM; lactate: 15 μM) [66] | Low LOD (enables pathogen/toxin detection at trace levels) [73] |
| Response Time | Seconds to minutes [66] | Rapid (real-time monitoring with SPR) [73] |
| Multiplexing Capability | Limited | Excellent (interferometric sensors enable simultaneous multi-analyte detection) [73] |
| Portability | High (miniaturization feasible, POC compatible) [74] | Varies (colorimetric assays highly portable; SPR systems less so) [73] |
| Sample Preparation | Minimal often required | May require extensive processing for complex matrices [73] |
| Cost | Low to moderate [74] | Moderate to high (depends on detection method) [73] |
| Resistance to Interference | Susceptible to electrode fouling, interferents in complex matrices [66] | Susceptible to background fluorescence, light scattering [73] |
Electrochemical biosensors typically offer superior sensitivity and lower detection limits, making them ideal for detecting low-abundance biomarkers in complex plant tissues [66]. Optical biosensors, particularly SPR and fluorescence-based platforms, provide exceptional specificity and real-time monitoring capabilities but may require more sophisticated instrumentation [73].
Applications in Agricultural Research
Electrochemical Biosensors in Agriculture
Electrochemical biosensors have demonstrated significant utility across various agricultural applications, particularly for monitoring plant health, detecting pathogens, and assessing food quality [72] [3]. Their portability, cost-effectiveness, and sensitivity make them well-suited for field-deployable diagnostics in precision agriculture.
Key application areas include:
- Plant Health Monitoring: Wearable and implantable electrochemical sensors enable real-time monitoring of phytohormones, stress biomarkers, and nutrient levels in plants [72]. For instance, Tang et al. (2024) demonstrated near-instantaneous detection of plant hormone fluctuations within minutes of pathogen exposure using differential pulse voltammetry [66]
- Pathogen Detection: Electrochemical biosensors facilitate early detection of devastating oilseed crop pathogens such as Sclerotinia sclerotiorum (stem rot) and Phakopsora pachyrhizi (soybean rust), enabling timely intervention before significant yield losses occur [3]
- Soil and Environmental Monitoring: These sensors can detect heavy metals, pesticides, and other environmental contaminants that affect crop health and food safety [77]
The integration of electrochemical biosensors with Internet of Things (IoT) platforms and artificial intelligence represents a promising direction for smart agriculture, enabling real-time, field-deployable disease surveillance and automated decision-support systems [72] [3].
Optical Biosensors in Agriculture
Optical biosensors have found diverse applications in food safety and quality assurance, leveraging their high sensitivity and specificity for detecting contaminants and pathogens throughout the food supply chain [73] [75].
Notable implementations include:
- Food Contaminant Detection: SPR biosensors effectively detect pesticide residues, toxins, and foodborne pathogens in agricultural products [73]. Fluorescence and chemiluminescence-based sensors offer amplified optical signals for detecting aflatoxins, mycotoxins, and other low-abundance analytes in complex food matrices [73]
- Plant Disease Diagnosis: Biomarker-targeted optical biosensors enable early diagnosis of plant diseases by detecting pathogen-specific proteins, nucleic acids, and metabolites [75]. For example, gold nanoparticle-based lateral flow biosensors have been developed for sensitive visual detection of potato late blight pathogen Phytophthora infestans [75]
- Quality Control: Optical biosensors monitor biochemical changes indicative of food freshness, storage conditions, and shelf life in processed and packaged food products [73]
Despite their impressive capabilities, optical biosensors face challenges related to non-specific interactions in complex food matrices, requiring advanced surface chemistry strategies and robust sensor coatings to maintain detection accuracy [73].
Experimental Protocols and Methodologies
Developing an Electrochemical Biosensor for Plant Pathogen Detection
Objective: Detect Sclerotinia sclerotiorum (stem rot pathogen) in oilseed crops using an electrochemical aptasensor [3].
Table 2: Research Reagent Solutions for Electrochemical Biosensing
| Reagent/Material | Function | Specification/Alternative |
|---|---|---|
| Aptamer probe | Biorecognition element | Specific to S. sclerotiorum DNA sequence |
| Gold nanoparticles | Electrode modification | 20-40 nm diameter, enhances surface area & electron transfer |
| Graphene oxide | Electrode substrate | Provides large surface area & excellent conductivity |
| Methylene blue | Redox indicator | Intercalates with dsDNA, generates electrochemical signal |
| Phosphate buffer | Electrolyte solution | 0.1 M, pH 7.4, provides optimal ionic strength |
| Nafion solution | Polymer binder | 0.5% wt, stabilizes biocomposite layer on electrode |
Procedure:
- Electrode Preparation: Modify a glassy carbon electrode (GCE) with graphene oxide-gold nanoparticle (GO-AuNP) nanocomposite to enhance surface area and electron transfer efficiency [3]
- Aptamer Immobilization: Covalently immobilize thiol-terminated aptamers specific to S. sclerotiorum DNA on the GO-AuNP/GCE using gold-thiol self-assembly [3]
- Hybridization and Detection:
- Incubate modified electrode with sample extract
- Perform electrochemical impedance spectroscopy (EIS) and differential pulse voltammetry (DPV) measurements
- Monitor current decrease or impedance increase due to DNA hybridization
- Signal Measurement: Use a portable potentiostat for field measurements. The sensor demonstrates a linear range of 1.0 fM to 10 nM with LOD of 0.3 fM [3]
Electrochemical Biosensor Workflow for Plant Pathogen Detection
Developing an Optical Biosensor for Mycotoxin Detection
Objective: Detect zearalenone (mycotoxin) in maize using a fluorescence-based biosensor [75].
Table 3: Research Reagent Solutions for Optical Biosensing
| Reagent/Material | Function | Specification/Alternative |
|---|---|---|
| Zearalenone aptamer | Biorecognition element | Specific to zearalenone mycotoxin |
| Cy5 fluorescent dye | Signal reporter | Covalently linked to aptamer |
| Gold nanoparticles | Fluorescence quencher | 20 nm diameter, quenches Cy5 emission |
| Quencher-labeled cDNA | Complementary DNA | Competes with target for aptamer binding |
| Tris-acetate buffer | Assay buffer | 20 mM, pH 7.4, 100 mM NaCl, 5 mM KCl |
| Microfluidic chip | Detection platform | Polydimethylsiloxane (PDMS) based |
Procedure:
- Probe Design: Synthesize a Cy5-labeled aptamer specific to zearalenone and complementary DNA (cDNA) functionalized with gold nanoparticles (AuNPs) as quenchers [75]
- Assay Principle: In absence of target, aptamer hybridizes with cDNA, bringing Cy5 close to AuNPs → fluorescence quenching. When zearalenone present, it binds aptamer, releasing cDNA → fluorescence recovery [75]
- Detection:
- Mix sample extract with sensing probe in tris-acetate buffer
- Incubate 15 minutes at room temperature
- Measure fluorescence intensity at 670 nm (excitation: 640 nm)
- Analysis: Sensor shows linear range 0.01-100 ng/mL with LOD of 0.003 ng/mL, below EU regulatory limits [75]
Optical Biosensor Workflow for Mycotoxin Detection
Current Challenges and Future Perspectives
Technical Limitations and Solutions
Both electrochemical and optical biosensors face significant challenges that impact their practical implementation in agricultural settings:
Electrochemical Biosensor Challenges:
- Biofouling and Electrode Degradation: Continuous exposure to complex plant matrices causes sensor performance deterioration [66]. Recent solutions include zwitterionic polymer coatings and epoxy propyl dimethyl ammonium chloride coatings that improve sensor stability for extended use [66]
- Limited Long-Term Stability: Achieving weeks to months of operational stability remains challenging. Strategies such as porous conductive polymers for enzyme entrapment and advanced immobilization techniques show promise for enhancing sensor lifespan [66]
- Miniaturization Limitations: Integrating multiple electrodes with different material requirements into compact form factors presents fabrication challenges [66]
Optical Biosensor Challenges:
- Matrix Interference: Non-specific interactions in complex food samples compromise detection accuracy. Advanced surface chemistry strategies and robust sensor coatings are being developed to address this limitation [73]
- Instrumentation Portability: While colorimetric assays are highly portable, more sophisticated optical platforms like SPR have limited field-deployability [73]
- Standardization and Validation: Many optical biosensors lack validated protocols and official approval for routine regulatory testing, hindering commercialization [73]
Emerging Trends and Future Directions
The future of biosensing in agricultural research will likely be shaped by several convergent technological trends:
- Integration with Digital Agriculture: Combining biosensors with IoT networks, drones, and AI-assisted data interpretation will enable real-time, field-deployable disease surveillance and automated decision-support systems [72] [3]
- Multiplexed Detection Platforms: Developing sensors capable of simultaneously monitoring multiple pathogens, stressors, or nutritional status indicators will provide comprehensive crop health assessment [66]
- Sustainable Sensor Materials: Research into biodegradable components and environmentally friendly fabrication processes will enhance the sustainability of biosensor technologies [3]
- Nanomaterial Innovations: Advanced nanomaterials including quantum dots, metal-organic frameworks, and graphene-based composites will continue to push detection limits and improve sensor performance [73] [3]
The global biosensors market reflects this growth trajectory, with the food and agriculture biosensors sector projected to expand from USD 7.7 billion in 2024 to USD 18.7 billion by 2034, representing a compound annual growth rate of 9.4% [77]. Similarly, the electrochemical biosensors market specifically is expected to grow from USD 7.32 billion in 2024 to approximately USD 16.45 billion by 2032 [78].
Electrochemical and optical biosensors offer complementary strengths for agricultural applications. Electrochemical platforms excel in sensitivity, portability, and cost-effectiveness, making them ideal for field-based monitoring of plant health markers and pathogens. Optical biosensors provide superior capabilities for multiplexed detection, real-time monitoring, and visual interpretation, advantageous for food safety and quality control applications.
The choice between these technologies ultimately depends on specific application requirements, including target analytes, detection limits, sample matrix complexity, and operational environment. Future advancements in nanomaterial integration, device miniaturization, and digital integration will further enhance the capabilities of both platforms, solidifying their role as indispensable tools for sustainable agriculture and food security.
The integration of electrochemical biosensors into agricultural research represents a paradigm shift towards rapid, on-site analysis for applications ranging from food safety to plant pathogen detection. For these novel sensing platforms to gain acceptance in research and regulatory contexts, they must demonstrate performance comparable to established, gold-standard laboratory methods. Validation is a critical process that establishes the reliability and credibility of new analytical techniques. This technical guide examines the framework for validating electrochemical biosensors against three cornerstone technologies: High-Performance Liquid Chromatography (HPLC) and Gas Chromatography-Mass Spectrometry (GC-MS) for contaminant and metabolite analysis, and Polymerase Chain Reaction (PCR) for pathogenic detection.
Gold-Standard Methods in Agricultural Analysis
Established analytical techniques provide the benchmark for sensitivity, specificity, and quantitative accuracy in agricultural science.
Chromatography and Mass Spectrometry (HPLC, GC-MS): These techniques separate complex mixtures and provide highly sensitive identification and quantification of chemical analytes. HPLC-UV/VIS is a well-established, targeted method for compounds like amino acids, often using inexpensive equipment available in most control laboratories [79]. GC-MS offers superior sensitivity and selectivity, capable of detecting and confirming the identity of compounds based on their mass fragmentation patterns [80]. It is widely used for a broad spectrum of metabolites, including sugars, organic acids, and amino acids [79]. Liquid Chromatography-Mass Spectrometry (LC-MS/MS) and High-Resolution Mass Spectrometry (HRMS) represent further advancements, providing high sensitivity and the capability to handle complex mixtures for the detection of contaminants like toxins and antibiotics [80] [81].
Polymerase Chain Reaction (PCR): This molecular technique amplifies specific DNA sequences, enabling the sensitive and specific detection of plant pathogens, such as fungi, bacteria, and viruses [82]. It is a fundamental tool for genosensors and the definitive method for confirming the presence of pathogenic DNA.
Limitations of Conventional Methods
Despite their accuracy, gold-standard methods present significant challenges for rapid, field-deployable agricultural monitoring.
- Resource Intensity: They often involve complex sample preparation, are time-consuming, and require expensive instrumentation [83] [81].
- Specialized Operation: These techniques demand highly trained technicians for operation and data interpretation, restricting their use to centralized laboratories [81].
- Limited Suitability for Field Use: Their inherent complexity makes them unsuitable for real-time, on-site decision-making in agricultural settings [17].
Validation Framework for Electrochemical Biosensors
Validation establishes the agreement between the new biosensor method and the reference method across key analytical performance metrics.
Key Analytical Performance Metrics
A comprehensive validation study should report and compare the following parameters against the gold standard:
Table 1: Key Performance Metrics for Biosensor Validation
| Performance Metric | Description | Comparison Goal |
|---|---|---|
| Limit of Detection (LOD) | The lowest concentration of an analyte that can be reliably detected. | Biosensor LOD should be comparable to or better than the application requires. |
| Linear Range | The concentration interval over which the sensor's response is linearly proportional to the analyte concentration. | Should cover the relevant physiological or contaminant concentration range. |
| Sensitivity | The slope of the analytical calibration curve, indicating the change in signal per unit change in concentration. | Should be sufficient to detect meaningful concentration changes. |
| Selectivity/Specificity | The sensor's ability to respond only to the target analyte in the presence of potential interferents. | Must demonstrate high specificity for the target in complex matrices (e.g., food, soil, sap). |
| Reproducibility | The precision of measurements under varied conditions (e.g., different operators, sensors, days), expressed as relative standard deviation (RSD). | RSD should typically be <5%, indicating robust manufacturing and protocol [84]. |
Experimental Design for Correlation Studies
The core of validation is a direct comparison using identical sample sets.
- Sample Preparation: A set of samples (e.g., contaminated food, infected plant tissue) is spiked with the target analyte across a range of concentrations relevant to the real-world application.
- Parallel Analysis: Each sample is split and analyzed simultaneously using the electrochemical biosensor and the reference method (e.g., HPLC, PCR).
- Data Correlation: Results from both methods are plotted against each other (biosensor response vs. reference method concentration), and statistical measures like the correlation coefficient (R²) and slope of the regression line are calculated. A strong correlation (e.g., R² > 0.98) indicates good agreement.
Table 2: Exemplary Performance of Validated Electrochemical Biosensors
| Target Analyte | Biosensor Type | Gold-Standard Comparison | Reported Biosensor Performance | Application Context |
|---|---|---|---|---|
| Pseudomonas aeruginosa (Bacteria) | Aptasensor | Culture-based methods (CFU counting) | LOD: 3.03 CFU mL⁻¹; High selectivity in food samples [84]. | Food Safety |
| Paracetamol (Drug) | MIP-based Sensor | High-Performance Liquid Chromatography (HPLC) | LOD: 10 nM; Excellent agreement with HPLC in pharmaceutical samples [84]. | Pharmaceutical Analysis |
| BRCA-1 Protein (Cancer Biomarker) | Immunosensor | Standard clinical diagnostics | LOD: 0.04 ng/mL; Linear Range: 0.05–20 ng/mL; Recovery in serum: 98 ± 3% [85]. | Medical Diagnostics |
| Amino Acids (e.g., Leucine) | Not Specified | HPLC-UV/VIS, GC-MS, NMR | Significantly higher levels detected in adulterated meat, concordant across all techniques [79]. | Food Authenticity |
Detailed Experimental Protocols
Protocol 1: Validation of a Biosensor for Chemical Contaminants Against HPLC
This protocol outlines the steps to validate an electrochemical biosensor for a pesticide against HPLC-UV.
Primary Materials:
- Electrochemical Biosensor: e.g., Screen-printed electrode (SPE) modified with a specific biorecognition element (aptamer, antibody, MIP).
- HPLC System: Equipped with a UV/VIS or diode-array detector (DAD).
- Analytical Standards: High-purity target analyte (e.g., specific pesticide) and internal standards.
- Mobile Phases: HPLC-grade solvents and buffers as required for the method.
- Supporting Electrolyte: Phosphate buffer saline (PBS) or other suitable buffer for electrochemical measurements.
Experimental Workflow:
- Calibration Curve Generation (HPLC):
- Prepare a series of standard solutions of the analyte at a minimum of five different concentrations.
- Inject each standard into the HPLC system in triplicate.
- Plot the peak area (or height) against concentration to establish the HPLC calibration curve, determining its LOD, LOQ, and linear range.
- Calibration Curve Generation (Biosensor):
- Under optimized conditions (pH, incubation time), measure the electrochemical response (e.g., via DPV or EIS) of the biosensor to the same standard concentrations.
- Plot the signal (e.g., current change, charge transfer resistance) against concentration to establish the biosensor's calibration curve.
- Analysis of Spiked Real Samples:
- Obtain a blank matrix (e.g., crop extract) and confirm the absence of the target analyte using HPLC.
- Spike the matrix with known concentrations of the analyte covering the low, medium, and high end of the linear range.
- Split each spiked sample and analyze using both the biosensor and the HPLC method.
- Data Analysis and Validation:
- Use a statistical test (e.g., Student's t-test) to confirm no significant difference between the results from the two methods.
- Perform linear regression analysis (biosensor result vs. HPLC result) to determine the correlation coefficient (R²), slope, and intercept.
- Calibration Curve Generation (HPLC):
Protocol 2: Validation of a Pathogen DNA Biosensor Against PCR
This protocol describes the validation of an electrochemical genosensor for a plant pathogen against PCR.
Primary Materials:
- Electrochemical Genosensor: e.g., Gold or carbon electrode modified with a single-stranded DNA (ssDNA) probe complementary to the target pathogen's DNA.
- PCR Thermocycler and Gel Electrophoresis Setup or qPCR system.
- DNA Extraction Kit: For purifying DNA from plant tissue or food samples.
- Hybridization Buffer: To facilitate specific binding between the probe and target DNA.
- Electrochemical Redox Marker: e.g., [Fe(CN)₆]³⁻/⁴⁻, to monitor the hybridization event via EIS or DPV.
Experimental Workflow:
- Sample Preparation and DNA Extraction:
- Inoculate plant material with the target pathogen (e.g., Sclerotinia sclerotiorum) and include healthy controls [15].
- Extract genomic DNA from all samples using the commercial kit.
- PCR Amplification and Analysis:
- Perform PCR using primers specific to the target pathogen's DNA.
- Analyze the PCR products using gel electrophoresis to confirm the presence/absence and size of the amplicon. For quantification, use qPCR.
- Biosensor Analysis:
- Hybridize the extracted DNA (or a specific PCR amplicon) with the probe on the genosensor surface.
- Wash the sensor to remove non-specifically bound DNA.
- Measure the electrochemical signal. A successful hybridization event typically causes a measurable change (e.g., an increase in charge transfer resistance in EIS).
- Data Comparison:
- Compare the biosensor's response (positive/negative, or quantitative signal) with the PCR results (presence/absence of a band, or Ct value from qPCR).
- Calculate the sensitivity (true positive rate) and specificity (true negative rate) of the biosensor against the PCR benchmark.
- Sample Preparation and DNA Extraction:
The Scientist's Toolkit: Essential Research Reagents and Materials
The development and validation of robust electrochemical biosensors rely on a suite of specialized reagents and materials.
Table 3: Key Research Reagent Solutions for Biosensor Development and Validation
| Reagent/Material | Function | Example in Context |
|---|---|---|
| Biorecognition Elements | Provides specificity by binding to the target analyte. | Aptamers ("chemical antibodies") for antibiotics [81]; Antibodies for pathogens like E. coli O157 [16]; DNA probes for pathogen DNA [83]. |
| Nanomaterials | Enhances electrode surface area and electron transfer, improving sensitivity. | Gold nanoparticles (AuNPs) for signal amplification in immunosensors [85]; Molybdenum disulfide (MoS₂) in nanocomposites [85]; Graphene-QD hybrids for femtomolar sensitivity [85]. |
| Electrode Materials | Serves as the physical transducer for the electrochemical signal. | Screen-printed electrodes (SPEs) for portable, cost-effective, disposable sensors [83]; Gold (Au) and Glassy Carbon (GC) for high-performance lab-based sensors [83]. |
| Electrochemical Redox Probes | Generates a measurable current signal that changes upon target binding. | [Fe(CN)₆]³⁻/⁴⁻ is commonly used to monitor impedance changes (in EIS) due to biomolecular binding on the electrode surface [83]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymers with cavities complementary to the target, serving as artificial receptors. | Used as a stable alternative to biological receptors for detecting small molecules like paracetamol or toxins [84] [83]. |
| Blocking Agents | Reduces non-specific binding on the sensor surface, improving selectivity. | Bovine Serum Albumin (BSA) is routinely used to block uncovered active sites on modified electrodes. |
Visualizing Workflows and Relationships
Biosensor Validation Workflow
The following diagram illustrates the multi-stage process of validating an electrochemical biosensor against a gold-standard method.
Method Comparison Landscape
This diagram provides a comparative overview of the operational characteristics of electrochemical biosensors versus traditional gold-standard methods.
The rigorous validation of electrochemical biosensors against gold-standard methods is not merely a procedural step but a fundamental requirement for their adoption in agricultural research and quality control. By systematically comparing performance metrics such as LOD, sensitivity, and specificity using structured experimental protocols, researchers can build a compelling case for the reliability of these novel tools. As biosensor technology continues to advance, overcoming challenges related to matrix interference and standardization, their validation will pave the way for a new era of precision agriculture. This will enable real-time, data-driven decision-making for enhanced food safety, plant pathogen management, and sustainable agricultural practices.
Electrochemical biosensors represent a transformative technology for agricultural research, offering the potential for rapid, on-site detection of pathogens, contaminants, and nutrients. While laboratory demonstrations of these sensors have shown exceptional sensitivity and selectivity, their translation from controlled research environments to real-world agricultural settings presents significant challenges. The environmental robustness of these analytical devices and their performance under actual field conditions constitute critical hurdles that must be overcome for widespread adoption. This technical assessment examines the key factors affecting field applicability, synthesizes current testing methodologies, and provides frameworks for evaluating electrochemical biosensors within authentic agricultural contexts, addressing a crucial gap between technological innovation and practical implementation [16] [17].
Agricultural environments present uniquely challenging conditions for biosensing platforms, including temperature fluctuations, variable humidity, chemical interferents, and complex sample matrices. Unlike clinical or industrial settings, agricultural applications often require operation in resource-limited environments without specialized technical expertise. Field testing under realistic conditions is therefore not merely a validation step but an essential component of the development process, revealing limitations often invisible in laboratory studies [16]. This review systematically addresses the primary challenges and evaluation methodologies necessary to advance electrochemical biosensors from promising prototypes to reliable field-deployable tools for precision agriculture and food safety monitoring.
Key Challenges in Field Deployment
Matrix Interference and Complex Samples
Agricultural samples, including soil extracts, plant sap, and food products, contain numerous compounds that can interfere with electrochemical sensing mechanisms. These complex matrices differ significantly from the buffer solutions typically used in laboratory development, presenting one of the most significant barriers to real-world implementation [16].
- Fouling and Non-specific Adsorption: Electrochemical sensors are particularly susceptible to surface fouling from proteins, polysaccharides, and organic matter present in agricultural samples. This fouling can significantly reduce sensitivity and selectivity by blocking active sites on the electrode surface and impeding electron transfer processes [24].
- Electrochemical Interferents: Compounds such as ascorbic acid, uric acid, and various phenolic compounds commonly found in plant and food samples can undergo redox reactions at similar potentials to target analytes, generating false positive signals and reducing measurement accuracy [16].
- Variable Sample Composition: Natural variations in pH, ionic strength, and viscosity across different sample types and environmental conditions can alter sensor performance, necessitating calibration protocols that account for this variability [83].
Environmental Stability and Sensor Lifespan
The operational stability of electrochemical biosensors under fluctuating environmental conditions remains a significant challenge for field deployment. Agricultural settings typically lack controlled temperature and humidity, creating demanding storage and operational requirements [16].
- Biorecognition Element Stability: Enzymes, antibodies, and aptamers can denature or degrade under field conditions, particularly when exposed to temperature extremes or repeated freeze-thaw cycles. This degradation directly impacts sensor reliability over time [16] [83].
- Nanomaterial Performance: While nanomaterials enhance sensitivity, their long-term stability in electrochemical sensors requires further investigation. Agglomeration, oxidation, or detachment from electrode surfaces can diminish the enhanced electrochemical properties that make them valuable for sensing applications [3].
- Packaging and Physical Integrity: Robust encapsulation that protects sensing elements from environmental damage while permitting sample access represents an ongoing engineering challenge, particularly for disposable, cost-effective sensors [16].
Usability and Integration Barriers
Beyond technical performance, practical considerations significantly influence the real-world applicability of electrochemical biosensors in agricultural settings [16].
- User-Friendly Operation: Successful field deployment requires interfaces and protocols accessible to agricultural professionals without specialized analytical expertise, including straightforward sample preparation, intuitive data interpretation, and minimal training requirements [16].
- System Integration: Standalone sensors offer limited utility compared to those integrated into broader decision-support systems. Connectivity with mobile platforms, data analytics, and agricultural IoT networks represents a critical value-enhancing capability [24] [16].
- Regulatory Standardization: The absence of standardized validation protocols and performance metrics specific to agricultural applications creates uncertainty regarding reliability and complicates comparative assessment between different sensor technologies [16] [83].
Table 1: Key Field Deployment Challenges and Impact on Sensor Performance
| Challenge Category | Specific Limitations | Impact on Sensor Performance |
|---|---|---|
| Sample Matrix Effects | Fouling, electrochemical interferents, variable pH/ionic strength | Reduced sensitivity and selectivity, false positives/negatives |
| Environmental Stability | Biorecognition element degradation, nanomaterial instability | Shortened operational lifespan, calibration drift |
| Usability & Integration | Complex sample preparation, limited connectivity, data interpretation | Limited adoption, reduced utility for decision support |
Methodologies for Field Testing and Validation
Environmental Stress Testing
Rigorous environmental testing establishes the operational boundaries and reliability of electrochemical biosensors under conditions mimicking real-world agricultural environments [16].
- Temperature Cycling: Expose sensors to temperature ranges reflecting expected field conditions (e.g., 5°C to 45°C) through multiple cycles while monitoring key performance parameters (sensitivity, response time, baseline stability) to assess thermal robustness [16].
- Humidity Resistance: Evaluate sensor performance and physical integrity under high humidity conditions (≥90% RH) typical of greenhouse environments or field use following precipitation events, with particular attention to electrical shorting and biofilm formation [15].
- Long-Term Stability Studies: Monitor calibration accuracy and signal output over extended periods (weeks to months) under both controlled storage conditions and intermittent operational use to establish expected field lifespan and recalibration requirements [16].
Real-Sample Analysis Protocols
Validating sensor performance with authentic agricultural samples provides critical data on matrix effects and practical utility [83].
- Parallel Analysis with Reference Methods: Compare biosensor results with established laboratory techniques (e.g., HPLC, GC-MS, ELISA) for identical sample sets to establish accuracy and reliability across diverse sample types and analyte concentrations [83] [50].
- Spike-and-Recovery Experiments: Fortify blank or reference matrices with known analyte concentrations across the expected detection range to quantify accuracy (percent recovery) and precision (coefficient of variation) in complex samples [83].
- Interference Testing: Methodically expose sensors to common agricultural interferents (e.g., soil humic acids, plant secondary metabolites, fertilizer components) at biologically relevant concentrations to quantify selectivity and identify potential cross-reactivities [16].
Performance Metrics for Field Applicability
Standardized metrics beyond conventional laboratory figures of merit are essential for evaluating real-world potential [16].
- Time-to-Result in Field Conditions: Measure the complete operational timeline from sample collection to result generation, including any necessary preparation steps, as this critically impacts practical utility for decision-making [24] [16].
- Operational Consistency: Quantify signal variance and detection limit fluctuations across different environmental conditions, lots of manufactured sensors, and multiple users to establish reliability expectations [16].
- User Experience Assessment: Document protocol complexity, training requirements, and interpretation challenges through structured trials with intended end-users (e.g., farmers, agricultural technicians) rather than experienced researchers [16].
Table 2: Essential Field Validation Experiments and Assessment Criteria
| Validation Protocol | Key Parameters Measured | Acceptance Criteria for Field Deployment |
|---|---|---|
| Environmental Stress Testing | Signal drift, physical integrity, recovery after stress | <15% signal deviation across operational temperature range |
| Real-Sample Analysis | Accuracy vs. reference methods, matrix effects | >80% recovery in fortified samples, strong correlation (R² > 0.95) with reference methods |
| End-User Trials | Success rate without training, time-to-result, interpretation accuracy | >90% successful operation by naive users, results in <30 minutes |
Strategies for Enhancing Environmental Robustness
Material Science Solutions
Advanced materials offer promising pathways to address the environmental challenges faced by electrochemical biosensors in agricultural settings [24] [3].
- Stabilized Biorecognition Elements: Employ immobilization matrices that preserve bioreceptor activity under field conditions. Hydrogel encapsulation, enzyme stabilization additives, and thermoresistant aptamer formulations can significantly enhance operational stability across temperature ranges [3].
- Robust Nanocomposites: Develop hybrid nanomaterials that maintain functionality under environmental stress. Metal-organic frameworks (MOFs) with protective cavities, graphene-polymer composites resistant to fouling, and ceramic-metallic hybrids offering improved temperature tolerance represent promising material approaches [3] [50].
- Anti-fouling Interfaces: Create electrode surfaces that resist non-specific adsorption through chemical modification. Polyethylene glycol (PEG) grafting, zwitterionic polymer coatings, and biomimetic surface topographies can significantly reduce matrix interference in complex agricultural samples [24].
Engineering and Design Approaches
Physical design and system architecture play crucial roles in determining field robustness and usability [16] [17].
- Microfluidic Sample Processing: Incorporate on-chip filtration, separation, or dilution capabilities to handle complex samples and reduce interferent concentration prior to detection, thereby minimizing matrix effects without requiring external equipment [24].
- Multi-Sensor Arrays: Employ sensor arrays with cross-reactive elements coupled with multivariate data analysis to differentiate target signals from interference patterns, enhancing effective selectivity in complex environments [16].
- Modular Design Principles: Develop systems with replaceable sampling modules or renewable sensing elements to address the fundamental limitation of bioreceptor degradation, effectively extending functional lifespan under challenging conditions [16].
Data Science and Signal Processing
Advanced data analysis techniques can extract reliable information from sensors operating in variable environmental conditions [16] [50].
- Environmental Compensation Algorithms: Integrate temperature, pH, and humidity sensors with correction algorithms that adjust electrochemical readings based on measured environmental parameters, significantly improving accuracy across field conditions [16].
- Machine Learning Classification: Train pattern recognition systems on comprehensive datasets collected under diverse environmental conditions and interfering substances to distinguish true analyte signals from background variations and matrix effects [16] [50].
- Drift Correction Methodologies: Implement self-referencing sensing schemes and periodic calibration checks with internal standards to identify and correct for signal drift over time, maintaining measurement reliability throughout sensor lifespan [16].
Environmental Robustness Strategies
Field Testing Experimental Framework
Tiered Validation Protocol
A systematic, multi-tiered approach to field testing ensures comprehensive evaluation while efficiently allocating resources. This progressive framework moves from controlled to fully authentic environments [16] [17].
- Tier 1: Simulated Field Testing - Conduct experiments in environmental chambers that replicate field conditions (temperature cycles, humidity variations) while using standardized samples. This controlled approach isolates environmental effects from matrix complications.
- Tier 2: Controlled Field Trials - Deploy sensors in operational agricultural settings but with researcher-managed protocols. Test with both spiked samples and selected authentic samples, comparing results with portable reference instruments.
- Tier 3: Authentic User Trials - Provide sensors to agricultural professionals (farmers, food processors, extension agents) for evaluation during normal operations. Monitor success rates, failure modes, and practical utility without researcher intervention.
Key Performance Indicators for Field Evaluation
Establishing quantitative metrics specific to field performance is essential for objective assessment and comparative analysis between different sensor technologies [16].
- Operational Reliability: Percentage of successful measurements obtained in field trials relative to total attempted measurements, with detailed documentation of failure modes and circumstances.
- Decision Concordance: Degree to which sensor-based decisions (e.g., apply pesticide, withhold harvest) align with those derived from reference laboratory methods, measuring practical impact rather than just analytical correlation.
- Total Operational Time: Complete time investment required per analysis, including sample collection, preparation, measurement, and data interpretation—often significantly longer than the measurement time alone reported in laboratory studies.
Table 3: Field Testing Protocol for Agricultural Biosensors
| Testing Phase | Sample Types | Validation Metrics | Success Criteria |
|---|---|---|---|
| Laboratory Benchmarking | Buffer standards, spiked samples | Sensitivity, selectivity, detection limit | Performance matching literature standards |
| Simulated Field Testing | Artificial matrices, temperature cycles | Signal stability, recovery rate | <20% performance degradation vs. controls |
| Controlled Field Trials | Authentic agricultural samples | Correlation with reference methods, false positive rate | >80% concordance with laboratory results |
| Authentic User Trials | Samples collected by end-users | Operational success rate, time-to-result | >85% successful operation by end-users |
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for Robust Biosensor Development
| Reagent Category | Specific Examples | Function in Sensor Development | Application Notes |
|---|---|---|---|
| Biorecognition Elements | Enzymes (glucose oxidase, HRP), antibodies, DNA aptamers | Target-specific molecular recognition | Select based on stability, not just affinity [83] |
| Signal Amplification Materials | Gold nanoparticles, graphene oxide, carbon nanotubes | Enhanced electrochemical signal | Improve sensitivity in complex matrices [3] [50] |
| Stabilization Matrices | Chitosan, Nafion, polyvinyl alcohol, hydrogels | Preserve bioreceptor activity | Critical for field deployment longevity [3] |
| Reference Materials | Certified analyte standards, artificial soil/plant matrices | Method validation and calibration | Essential for quantifying matrix effects [83] |
Field Testing Framework
The transition of electrochemical biosensors from research laboratories to agricultural fields necessitates rigorous attention to environmental robustness and comprehensive field validation. While significant progress has been made in sensor sensitivity and selectivity under controlled conditions, addressing the challenges of real-world deployment requires integrated solutions spanning materials science, engineering design, and data analytics. The experimental frameworks and assessment metrics outlined in this review provide structured approaches for evaluating and enhancing field applicability. Future advancements will likely focus on intelligent sensor systems that automatically compensate for environmental variables, robust manufacturing that ensures consistent performance across production batches, and seamless integration with agricultural decision-support platforms. Through systematic attention to field testing and environmental robustness, electrochemical biosensors can realize their potential to transform agricultural monitoring, enabling precision agriculture practices and enhancing global food security.
Electrochemical biosensors represent a transformative technology for the agricultural sector, offering a paradigm shift from centralized laboratory testing to decentralized, on-site analysis. Framed within the broader thesis of introducing electrochemical biosensors into agricultural research, this technical guide provides a critical examination of the economic and operational factors governing their field deployment. These analytical devices combine a biological recognition element with an electrochemical transducer, converting specific biochemical reactions into quantifiable electrical signals such as voltage, current, or impedance [25]. For researchers and scientists driving innovation in agri-food applications, understanding the cost structures, analytical performance capabilities, and usability parameters is essential for transitioning these technologies from laboratory prototypes to practical field solutions. This analysis focuses specifically on their application for detecting pathogens, toxins, and pesticides directly in field settings, processing facilities, and throughout the food supply chain, addressing the critical need for rapid intervention and data-driven decision support in contemporary agriculture [16].
Economic Analysis: Cost Structures and Market Projections
The economic viability of electrochemical biosensors is a primary determinant of their adoption potential. A comprehensive understanding of both the current market landscape and the underlying cost components is crucial for research planning and technology development.
Global Market Outlook
The biosensors market is experiencing significant growth, driven by demand across healthcare, environmental monitoring, and agri-food sectors. Table 1 summarizes the key market projections, highlighting the substantial market share held by electrochemical technologies and the promising growth in the food and agriculture segment [86].
Table 1: Global Biosensors Market Overview and Projections
| Metric | Value | Time Period/Notes |
|---|---|---|
| Total Biosensors Market Size | USD 30.25 Billion | 2024 [86] |
| Projected Market Size | USD 69.67 Billion | 2034 [86] |
| Compound Annual Growth Rate (CAGR) | 8.7% | 2025-2034 [86] |
| Food & Agriculture Biosensors Market Size | USD 7.7 Billion | 2024 [87] |
| Projected Food & Agriculture Market Size | USD 18.7 Billion | 2034 [87] |
| Food & Agriculture CAGR | 9.4% | 2025-2034 [87] |
| Dominant Technology Segment | Electrochemical Biosensors | Low cost, high specificity, and scalability [86] |
The data indicates that the food and agriculture segment is growing at a marginally faster rate than the overall biosensors market, signaling increasing investment and application potential in this sector [87] [86]. North America currently holds the largest market share (>41%), a position attributed to its advanced healthcare infrastructure, R&D investment, and progressive regulatory stance. However, the Asia-Pacific region is anticipated to witness the most rapid growth, driven by its large population, expanding agricultural and food industries, and increasing government initiatives in digital health and food safety [86].
Cost-Benefit Considerations for On-Site Deployment
The economic advantage of electrochemical biosensors is most apparent when evaluating their operational deployment. The core cost benefits stem from several key factors:
- Low-Cost Engineering and Miniaturization: The production of electrochemical biosensors is amenable to scaling using low-cost manufacturing techniques such as screen printing and inkjet printing, which produce disposable, single-use electrodes. This significantly reduces per-test costs [25].
- Reduced Reliance on Centralized Labs: On-site detection eliminates the logistical expenses and time delays associated with transporting samples to a central laboratory, which often requires cold-chain logistics and involves significant labor costs [16].
- Minimal Reagent and Sample Consumption: The integration with microfluidic Lab-on-a-Chip (LoC) platforms allows complex analyses to be performed with very small volumes of samples and reagents, reducing consumption and cost per test [25] [24].
- Prevention of Economic Losses: In agricultural applications, the early detection of crop pathogens like Sclerotinia sclerotiorum or Phakopsora pachyrhizi can prevent yield losses of 10-80%, representing savings of billions of dollars globally [3] [23]. Similarly, in food safety, rapid detection of toxins and pathogens can prevent costly product recalls and protect brand reputation.
Operational Analysis: Speed and Performance Metrics
The operational superiority of electrochemical biosensors for on-site use is demonstrated through their rapid analysis times and high sensitivity, outperforming traditional methods.
Performance Comparison with Conventional Techniques
Table 2 provides a comparative analysis of electrochemical biosensors against gold-standard laboratory methods, highlighting the transformative gains in speed and portability that enable on-site decision-making.
Table 2: Performance Comparison: Electrochemical Biosensors vs. Conventional Methods
| Parameter | Electrochemical Biosensors | Traditional Methods (PCR, ELISA) |
|---|---|---|
| Analysis Time | Minutes to a few hours [7] [16] | Several hours to days (including culture steps) [7] |
| Portability | High (portable, handheld, smartphone-integrated) [25] [24] | Low (requires fixed-laboratory setup) |
| Sensitivity | High (enabled by nanomaterials; e.g., pathogen detection at pico- to femtomolar levels) [25] [23] | High |
| On-Site/Field Use | Yes (designed for point-of-care testing) [16] [25] | No |
| Sample Preparation | Minimal, often integrated into LoC systems [25] [24] | Extensive, multi-step processes required |
| Multiplexing Capability | Emerging (capable of simultaneous detection of multiple analytes) [23] | Typically limited to single-analyte detection |
The significant reduction in analysis time—from days to minutes—is a critical operational advantage, allowing for immediate intervention in field and processing environments [16]. For instance, the early detection of soybean rust pathogens can facilitate timely fungicide application before the disease causes irreversible damage to photosynthesis, potentially saving millions in yield losses [3].
Key Performance Metrics and Technological Enablers
The enhanced performance of modern electrochemical biosensors is driven by key technological innovations:
- Nanomaterial Integration: The use of nanomaterials such as gold nanoparticles (AuNPs), graphene oxide (GO), carbon nanotubes, and metal-organic frameworks (MOFs) is pivotal. These materials enhance sensor sensitivity by providing a high surface-to-volume ratio for bioreceptor immobilization, improving electron transfer kinetics, and enabling signal amplification. This allows for the detection of targets at ultra-low concentrations (e.g., pathogens, toxins) directly in complex matrices [25] [3] [23].
- Limit of Detection (LOD): Nanomaterial-enhanced biosensors have achieved LODs sufficient for practical applications, such as detecting viral particles, bacterial cells, or mycotoxins at clinically and agriculturally relevant levels, often surpassing the sensitivity of traditional methods in a fraction of the time [23].
- Signal Transduction Mechanisms: The primary electrochemical techniques employed are:
- Amperometry: Measures current resulting from redox reactions at a constant potential.
- Voltammetry (e.g., Cyclic, Differential Pulse): Measures current while varying the applied potential.
- Impedance Spectroscopy: Measures the opposition to current flow, useful for label-free detection of binding events [25] [23].
- Recognition Elements: The specificity of these sensors is conferred by various biorecognition elements, including enzymes, antibodies, aptamers (synthetic single-stranded DNA/RNA), nucleic acids, and molecularly imprinted polymers (MIPs), each selected based on the target analyte and application requirements [25] [24].
User-Friendliness and Integration for Field Deployment
For a technology to be successfully adopted for on-site use, it must overcome barriers related to usability, stability, and integration into existing workflows.
Enhancing Usability and Connectivity
Recent advancements have focused on creating intuitive, connected systems that are accessible to non-specialists:
- Smartphone Integration: The coupling of electrochemical biosensors with smartphones is a cornerstone of user-friendly design. Smartphones provide a ubiquitous platform for powering the sensor, processing data, displaying intuitive results, and connecting to cloud storage via wireless networks. This transforms the system into a mobile laboratory, enabling real-time data sharing and decision support [25] [24].
- Lab-on-a-Chip (LoC) and Microfluidics: LoC systems integrate multiple laboratory functions—such as sample preparation, reaction, separation, and detection—onto a single, miniaturized chip. This automation simplifies the user's workflow to a "sample-in, answer-out" process, minimizing the need for technical expertise and reducing the risk of user error [25].
- Artificial Intelligence (AI) and Machine Learning (ML): The integration of AI/ML algorithms assists with data interpretation, enhances sensor calibration, compensates for matrix effects, and can even predict contamination risks or disease outbreaks based on real-time data streams, providing a powerful decision-support tool for farmers and quality control inspectors [7] [16] [23].
Operational Workflow for On-Site Analysis
The following diagram illustrates the streamlined, end-to-end workflow for conducting on-site analysis with an integrated electrochemical biosensing system.
Persistent Challenges and Mitigation Strategies
Despite the promising advancements, several challenges hinder widespread field deployment, alongside the research efforts to address them:
- Real-World Validation: A critical systematic review highlighted that only 1 out of 77 studies conducted validation on naturally contaminated food samples, with the majority relying on artificially spiked samples. This raises concerns about real-world reliability [7].
- Sensor Stability and Biofouling: Biological recognition elements (e.g., enzymes, antibodies) can degrade over time, and sensor surfaces are susceptible to fouling from non-specific adsorption in complex samples, leading to signal drift and reduced lifespan [86].
- Standardization and Regulatory Hurdles: A lack of standardized fabrication protocols, calibration methods, and validation procedures limits comparability between studies and complicates regulatory approval [7] [16].
The Scientist's Toolkit: Essential Research Reagents and Materials
The development and operation of high-performance electrochemical biosensors rely on a suite of specialized materials and reagents. Table 3 details the key components and their functions, forming an essential toolkit for researchers in this field.
Table 3: Research Reagent Solutions for Electrochemical Biosensor Development
| Material/Reagent | Function | Application Example |
|---|---|---|
| Gold Nanoparticles (AuNPs) | Enhance conductivity; provide large surface area for bioreceptor immobilization; catalytic properties [25]. | Signal amplification in pathogen [7] and virus detection [23]. |
| Graphene Oxide (GO) & Carbon Nanotubes (CNTs) | High surface area scaffold; excellent electrical conductivity; facilitates electron transfer [25] [23]. | Electrode modification for sensitive detection of toxins and pesticides [16]. |
| Aptamers | Synthetic single-stranded DNA/RNA recognition elements; high stability, affinity, and specificity to target [25] [24]. | Alternative to antibodies for detecting pathogens (e.g., Salmonella) [7] or small molecules [16]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymer with tailor-made cavities for specific target molecules; highly stable [25] [24]. | Detection of pesticides, antibiotics, or toxins in complex food samples [25]. |
| Antibodies | Biological recognition elements providing high specificity and affinity to antigens [25]. | Immunosensors for pathogen detection (e.g., E. coli O157, Listeria) [7] [16]. |
| Lab-on-a-Chip (LoC) Substrates | Miniaturized platforms (e.g., PDMS, PMMA) that integrate and automate all analytical steps [25] [24]. | Portable, "sample-in-answer-out" devices for on-site testing of food contaminants [25]. |
Detailed Experimental Protocols
To ensure reproducibility and provide a clear technical roadmap, this section outlines generalized, yet detailed, methodologies for key application areas.
Protocol for Pathogen Detection (e.g.,Salmonella spp.,E. coli)
This protocol is adapted from methodologies reviewed in systematic assessments of electrochemical biosensors for foodborne pathogens [7].
Biosensor Functionalization:
- Electrode Preparation: Clean the working electrode (e.g., screen-printed carbon or gold electrode) sequentially with ethanol and deionized water. Dry under a nitrogen stream.
- Nanomaterial Modification: Drop-cast a suspension of multi-walled carbon nanotubes (MWCNTs) or graphene oxide (GO) onto the electrode surface and allow to dry. This enhances the electroactive surface area.
- Bioreceptor Immobilization: Immobilize specific capture probes onto the modified electrode. For an aptasensor, incubate with a thiol- or amino-terminated aptamer specific to the target pathogen, followed by blocking with mercaptohexanol or BSA to minimize non-specific binding. For an immunosensor, immobilize a capture antibody using a cross-linker like EDC/NHS.
Sample Preparation and Assay:
- Sample Processing: For a food sample (e.g., 25g of leafy greens), homogenize in a buffered peptone water enrichment broth. Incubate for a short pre-enrichment period (if necessary). Centrifuge and filter the sample to obtain a supernatant.
- Incubation: Incubate the prepared sample supernatant on the functionalized biosensor for a defined period (e.g., 15-30 minutes) to allow pathogen binding.
- Electrochemical Measurement: Wash the sensor to remove unbound material. Perform electrochemical measurement in a suitable redox solution (e.g., containing [Fe(CN)₆]³⁻/⁴⁻). Use Electrochemical Impedance Spectroscopy (EIS) to measure the increase in charge transfer resistance (Rₑₜ) proportional to pathogen concentration.
Data Analysis:
- Calibrate the sensor with standards of known concentration.
- Plot the change in signal (e.g., Rₑₜ) against the logarithm of pathogen concentration to generate a calibration curve.
- Use this curve to interpolate the concentration of the target pathogen in unknown samples.
Protocol for Pesticide Residue Detection
This protocol is based on enzymatic inhibition principles and nanomaterial-enhanced signal transduction, as described in reviews on agri-food monitoring [16] [25].
Enzyme-Based Biosensor Fabrication:
- Electrode Modification: Deposit a nanocomposite of gold nanoparticles and reduced graphene oxide (AuNP-rGO) on the electrode to create a highly conductive and catalytic platform.
- Enzyme Immobilization: Co-immobilize the enzyme acetylcholinesterase (AChE) and the electron mediator methylene blue onto the modified electrode using a biopolymer like chitosan or Nafion to form a stable film.
Measurement Procedure (Amperometry):
- Baseline Current Measurement: Place the biosensor in a buffer solution and apply a fixed potential. Measure the baseline amperometric current resulting from the enzymatic conversion of a substrate (e.g., acetylthiocholine) into an electroactive product.
- Inhibition Step: Incubate the biosensor with the extracted sample solution containing the pesticide (e.g., organophosphate) for a fixed time (e.g., 10 minutes). Pesticides will inhibit the AChE enzyme.
- Post-Inhibition Measurement: Re-measure the amperometric current in the substrate solution. The degree of current reduction is proportional to the concentration of the inhibiting pesticide in the sample.
Quantification:
- Calculate the percentage of enzyme inhibition: % Inhibition = [(I₀ - Iₛ)/I₀] × 100, where I₀ is the initial current and Iₛ is the current after sample incubation.
- Quantify the pesticide concentration by referring to a calibration curve of % Inhibition vs. pesticide concentration.
Electrochemical biosensors present a compelling solution for on-site monitoring in agriculture and food safety, offering a powerful combination of cost-effectiveness, rapid analysis, and growing user-friendliness. The economic analysis confirms a robust and expanding market, underpinned by compelling operational advantages that enable a shift from reactive to proactive management of agricultural threats and food contaminants. However, the path to widespread adoption requires the research community to address persistent gaps, most notably the lack of real-world validation and the need for standardized protocols. The ongoing integration of these sensors with smartphone technology, AI-driven analytics, and IoT networks is poised to create intelligent, decision-support systems that transcend simple detection tools. For researchers and scientists, the future lies in developing these integrated, robust, and truly field-deployable systems that are not only sensitive and specific but also stable, reproducible, and validated under real-world conditions of use.
Conclusion
Electrochemical biosensors represent a paradigm shift in agricultural monitoring, moving from reactive to proactive crop management. Their core strengths lie in exceptional sensitivity, portability, and capacity for real-time, on-site analysis, which are critical for the early detection of pathogens and stressors. The integration of nanotechnology and smart materials has been pivotal in enhancing sensor performance, enabling the detection of targets at ultra-low concentrations. However, the journey from laboratory proof-of-concept to widespread field adoption requires overcoming significant hurdles related to sensor stability in complex matrices, standardization of protocols, and seamless integration into agricultural IoT networks. Future advancements will likely be driven by the development of multi-analyte detection platforms, AI-powered data interpretation, biodegradable sensor materials, and robust smartphone-integrated systems. For researchers and drug development professionals, the underlying biosensing principles and innovation in biorecognition elements offer valuable cross-disciplinary insights, paving the way for next-generation diagnostic tools that strengthen the entire biotechnological value chain, from sustainable agriculture to personalized medicine.
References
- 1. Bioreceptors as the key components for electrochemical ... [https://www.sciencedirect.com/science/article/pii/S2666386424000213]
- 2. Fundamentals of bio-electrochemical sensing [https://www.sciencedirect.com/science/article/pii/S266682112300073X]
- 3. Electrochemical Biosensors for Oilseed Crops [https://www.mdpi.com/2304-8158/14/16/2881]
- 4. Electrochemical Sensors for Sustainable Precision Agriculture ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9124781/]
- 5. Electrochemical Biosensors - Sensor Principles and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3663003/]
- 6. What are Electrochemical Biosensors? [https://www.news-medical.net/life-sciences/What-are-Electrochemical-Biosensors.aspx]
- 7. A systematic review of electrochemical biosensors for ... [https://www.sciencedirect.com/science/article/abs/pii/S0956713525004724]
- 8. Biosensors, Volume 15, Issue 8 (August 2025) – 85 articles [https://www.mdpi.com/2079-6374/15/8]
- 9. An electrochemical biosensor for detecting pathogenic ... [https://pubmed.ncbi.nlm.nih.gov/40749503/]
- 10. Recent Advances in Electrochemical Biosensors for ... [https://www.mdpi.com/journal/chemosensors/special_issues/RAEBABEA]
- 11. Advances in Impedimetric Biosensors: Current Applications ... [https://www.mdpi.com/2072-666X/16/11/1244]
- 12. Advances in Electrochemical Sensors for Monitoring of ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993625003711]
- 13. Real-Time, In Situ Detection for Precision Agriculture [https://www.sciencedirect.com/science/article/abs/pii/S0039914025014407]
- 14. Electrochemical biosensors: The beacon for food safety ... [https://www.sciencedirect.com/science/article/pii/S0308814625005357]
- 15. Electrochemical Biosensors for Oilseed Crops [https://pmc.ncbi.nlm.nih.gov/articles/PMC12385818/]
- 16. Electrochemical Biosensors for Smart Agri-Food Monitoring ... [https://www.sciencedirect.com/science/article/abs/pii/S2451910325001292]
- 17. Electrochemical Sensors for Sustainable Precision ... [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2022.848320/full]
- 18. Biosensors Metrics Glossary of Definitions - Zimmer & Peacock [https://www.zimmerpeacocktech.com/knowledge-base/faq/biosensors-glossary/]
- 19. An overview of sensitivity and selectivity of biosensors for ... [https://www.sciencedirect.com/science/article/abs/pii/B9780128146798000030]
- 20. On the Determination of Uncertainty and Limit of Detection ... [https://www.mdpi.com/1424-8220/18/7/2038]
- 21. Biosensors for Monitoring, Detecting, and Tracking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11545827/]
- 22. Future perspectives of GMO detection in agriculture [https://link.springer.com/article/10.1007/s00604-025-07267-x]
- 23. Rapid and accurate detection of crop viruses by nano ... [https://pubs.rsc.org/en/content/articlehtml/2025/ay/d5ay01205h]
- 24. Smartphone-Integrated Electrochemical Devices for ... [https://www.mdpi.com/2079-6374/15/9/574]
- 25. Smartphone-Integrated Electrochemical Devices for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467198/]
- 26. Recent Progress on Techniques in the Detection of Aflatoxin B1 in... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9573432/]
- 27. contamination in food crops: causes, Aflatoxin , and... detection [https://fppn.biomedcentral.com/articles/10.1186/s43014-021-00064-y]
- 28. A disposable paper-based electrochemical biosensor for ... [https://www.sciencedirect.com/science/article/pii/S0023643824012301]
- 29. Bio-nanoparticles sensor couple with smartphone digital ... [https://www.nature.com/articles/s41598-025-92944-3]
- 30. Evaluating Sap Analysis: A New Tool for Nutrient ... [https://cropaia.com/blog/sap-analysis/]
- 31. Electrochemical sensors for plant signaling molecules [https://www.sciencedirect.com/science/article/abs/pii/S0956566324007632]
- 32. Electrochemical sensors for plant signaling molecules [https://pubmed.ncbi.nlm.nih.gov/39250871/]
- 33. Plant Sap Analysis | Newage Laboratories [https://www.newagelaboratories.com/plant-sap-analysis]
- 34. Optimizing Whole-Cell Biosensors for the Early Detection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12109988/]
- 35. Smart and sustainable nano-biosensing technologies for ... [https://www.sciencedirect.com/science/article/pii/S0926669025002596]
- 36. Comprehensive review of emerging contaminants [https://www.sciencedirect.com/science/article/pii/S0147651324004962]
- 37. Advances in Detection Technologies for Pesticide Residues ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11941943/]
- 38. Electrochemical Biosensors: A Solution to Pollution ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6023016/]
- 39. A Strategy to Limit the Spread of Antibiotic-Resistant Bacteria [https://www.mdpi.com/2227-9040/13/9/352]
- 40. Development of CPE/ssDNA-Based Electrochemical ... [https://www.mdpi.com/2079-6374/15/11/708]
- 41. Integration of smart sensors and IOT in precision agriculture [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1587869/full]
- 42. AI-Enhanced Electrochemical Sensing Systems: A Paradigm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467342/]
- 43. Application of artificial intelligence in electrochemical ... [https://link.springer.com/article/10.1007/s44373-025-00042-w]
- 44. Smart farming for a sustainable future: implementing IoT ... [https://bnrc.springeropen.com/articles/10.1186/s42269-025-01366-8]
- 45. Artificial intelligence−powered electrochemical sensor [https://www.sciencedirect.com/science/article/pii/S2405844024139953]
- 46. Internet of Things and smart sensors in agriculture: Scopes ... [https://www.sciencedirect.com/science/article/pii/S2666154323002831]
- 47. Soil microbiome characterization and its future directions with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11389470/]
- 48. Matrix Effect Study and Immunoassay Detection Using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6187333/]
- 49. A review of biosensor for environmental monitoring [https://pmc.ncbi.nlm.nih.gov/articles/PMC10308875/]
- 50. Electrochemical Biosensors Driving Model Transformation ... [https://www.mdpi.com/2304-8158/14/15/2669]
- 51. A paper biosensor for overcoming matrix effects interfering ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10579119/]
- 52. Compensate for or Minimize Matrix Effects? Strategies ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7412464/]
- 53. A Look at Matrix Effects [https://www.chromatographyonline.com/view/look-matrix-effects]
- 54. Recent advances in nano-enhanced biosensors [https://www.sciencedirect.com/science/article/pii/S2214180425000492]
- 55. Nanomaterial-Mediated Electrochemical and Optical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12473853/]
- 56. Innovations in graphene-based electrochemical biosensors in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11982133/]
- 57. Graphene-based functional electrochemical sensors for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10894149/]
- 58. Gold Nanoparticles dotted Reduction Graphene Oxide ... [https://www.nature.com/articles/s41598-017-05352-7]
- 59. Signal amplification strategies in electrochemical ... [https://pubs.rsc.org/en/content/articlelanding/2022/ma/d2ma00427e]
- 60. A highly sensitive electrochemical sensor based on poly(3- ... [https://www.sciencedirect.com/science/article/abs/pii/S1001074223004552]
- 61. Progress on nanomaterials based-signal amplification ... [https://www.sciencedirect.com/science/article/pii/S2590137021000200]
- 62. Implantable electrochemical biosensors: Challenges, ... [https://www.sciencedirect.com/science/article/pii/S2451910325001048]
- 63. Wyss Institute develops coating to extend lifespan of ... [https://www.medicaldesignandoutsourcing.com/wyss-institute-researchers-develop-coating-to-extend-lifespan-of-implantable-biosensors/]
- 64. A target-triggering strategy for self-powered biosensor ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400525001364]
- 65. Polymer-Based Electrochemical Sensors for the Diagnosis of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12106273/]
- 66. Recent Developments in Microneedle Biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12388307/]
- 67. Device integration of electrochemical biosensors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9951169/]
- 68. A Cost-Effective and Rapid Manufacturing Approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11944730/]
- 69. Different approaches for fabrication of low-cost ... [https://www.sciencedirect.com/science/article/pii/S2451910321002076]
- 70. Biosensors for Sustainable Food Engineering: Challenges ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5872071/]
- 71. Electrochemical vs. Optical Biosensors for Point-of-Care ... [https://www.mdpi.com/2227-9040/11/10/546]
- 72. Recent advances in wearable and implantable ... [https://www.sciencedirect.com/science/article/pii/S0165993625002043]
- 73. Advancements in Optical Biosensor Technology for Food ... [https://www.mdpi.com/2673-4591/106/1/6]
- 74. Point-of-care biosensors for infectious disease diagnosis [https://pubs.rsc.org/en/content/articlehtml/2025/ra/d5ra03897a?page=search]
- 75. Research progress in biomarker-targeted optical ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993624003248]
- 76. An Overview of Optical and Electrochemical Sensors and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7915219/]
- 77. Food and Agriculture Biosensors Market Fueled by High ... [https://www.openpr.com/news/4022767/food-and-agriculture-biosensors-market-fueled-by-high-demand]
- 78. Electrochemical-Based Biosensors Market by Application [https://www.linkedin.com/pulse/electrochemical-based-biosensors-market-application-ihqqc/]
- 79. Comparison of Targeted (HPLC) and Nontargeted (GC-MS ... [https://www.mdpi.com/2304-8158/9/8/1084]
- 80. Recent Advances in the Detection of Food Toxins Using Mass ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10731662/]
- 81. revolutionizing antibiotic detection with aptamer-based ... [https://pubs.rsc.org/en/content/articlehtml/2025/ra/d5ra02100f?page=search]
- 82. PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12141547/]
- 83. Electrochemical (Bio)Sensors for Toxins, Foodborne ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12293878/]
- 84. Electrochemical Biosensor Applications in Agriculture, ... [https://www.mdpi.com/journal/biosensors/special_issues/9R091J1GJD]
- 85. Biosensors, Volume 15, Issue 5 (May 2025) – 69 articles [https://www.mdpi.com/2079-6374/15/5]
- 86. Biosensors Market | USD 69.67 Billion by 2034 [https://www.novaoneadvisor.com/report/biosensors-market]
- 87. Food & Agriculture Biosensors Market Share, Size, Growth ... [https://www.insightaceanalytic.com/report/global-food--agriculture-biosensors-market/1302]