Nanomaterial-Modified Screen-Printed Electrodes for Advanced Pesticide Analysis: From Fundamentals to Biomedical Applications
This comprehensive review explores the cutting-edge development and application of nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, addressing critical needs in food safety, environmental monitoring, and biomedical research.
Nanomaterial-Modified Screen-Printed Electrodes for Advanced Pesticide Analysis: From Fundamentals to Biomedical Applications
Abstract
This comprehensive review explores the cutting-edge development and application of nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, addressing critical needs in food safety, environmental monitoring, and biomedical research. The article systematically examines the foundational principles of SPE design and nanomaterial enhancement, detailed methodologies for electrode modification and pesticide detection, optimization strategies for improving sensor performance, and rigorous validation against conventional analytical techniques. Tailored for researchers, scientists, and drug development professionals, this work highlights how these portable, cost-effective biosensors enable rapid, sensitive, and selective detection of various pesticide classes including organophosphates, carbamates, and neonicotinoids, with significant implications for public health protection and clinical diagnostics.
Fundamentals of SPEs and Nanomaterial Enhancement for Pesticide Sensing
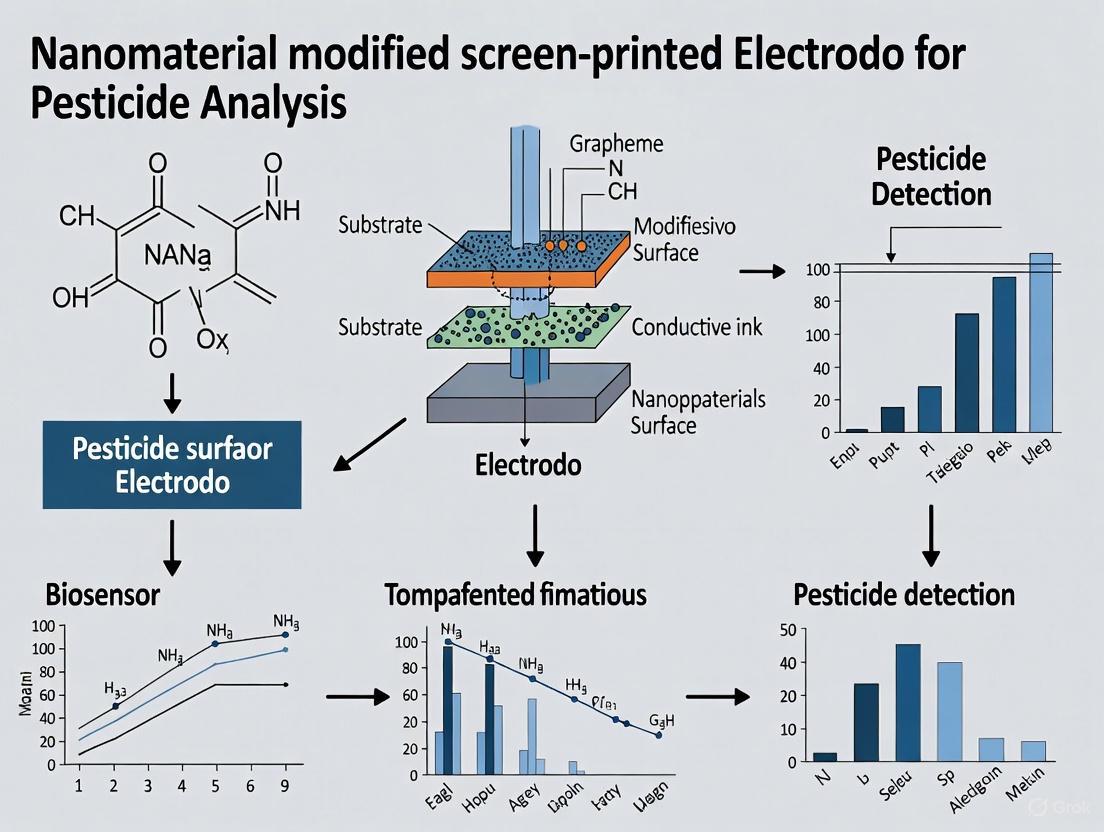
Screen-printed electrodes (SPEs) represent a transformative technology in electrochemistry, enabling the mass production of disposable, cost-effective, and portable sensing platforms. These miniaturized electrochemical cells have become fundamental tools for decentralized analysis across numerous fields, including environmental monitoring, clinical diagnostics, and food safety [1] [2]. Their significance is particularly pronounced in the context of pesticide analysis, where on-site detection capabilities offer a compelling alternative to traditional laboratory-based methods [1] [3]. SPEs integrate working, counter, and reference electrodes onto a single, compact substrate through a scalable printing process, making sophisticated electrochemical analysis accessible outside centralized laboratories [4] [2]. This application note details the design principles, fabrication methodologies, and inherent advantages of SPEs, with specific consideration to their application in pesticide detection research utilizing nanomaterial modifications.
Design and Structural Features of Screen-Printed Electrodes
The architecture of a typical screen-printed electrode is designed for functional completeness and miniaturization. A standard SPE comprises three primary components printed on a single, non-conductive substrate: a working electrode (WE), a counter electrode (CE), and a reference electrode (RE) [4] [2]. This integrated design creates a full electrochemical cell that is both compact and ready-to-use.
- Substrate Materials: The foundation of an SPE is a non-conductive material that provides mechanical stability. Common substrates include polyvinyl chloride (PVC), polyester, ceramics, and polycarbonate [5] [4]. The choice of substrate influences flexibility, chemical resistance, and overall durability.
- Conductive Inks: The electrode components are formed using conductive inks. The working electrode is often fabricated from carbon-based inks (e.g., graphite, graphene, carbon nanotubes) due to their wide potential window, chemical stability, and low cost [4]. The reference electrode is commonly made from silver/silver chloride (Ag/AgCl) ink, while the counter electrode is typically carbon or sometimes platinum [4].
- Design Flexibility: SPE designs are highly customizable. Using software like CorelDraw, researchers can create specific templates to guide the printing process, allowing for customization of electrode size, geometry, and layout to suit particular experimental or device integration needs [4].
The following diagram illustrates the typical layered structure and components of a standard screen-printed electrode.
Fabrication Process
The fabrication of SPEs is a multi-step, additive manufacturing process that allows for high-volume production. The process is valued for its simplicity, cost-effectiveness, and versatility [4] [2].
- Ink Preparation: The process begins with the formulation of conductive inks. These are viscous pastes composed of a conductive material (e.g., graphite, silver), a binder to control viscosity and adhesion, and organic solvents to create a homogeneous mixture [4]. The ink's rheological properties are critical for successful printing.
- Screen Printing: A mesh screen, patterned with the desired electrode design, is placed over the substrate. Conductive ink is forced through the open areas of the mesh onto the substrate using a squeegee. This step is performed sequentially for each ink type (e.g., carbon for WE/CE, then Ag/AgCl for RE) [5] [4].
- Curing and Drying: After printing, the electrodes are cured in an oven to evaporate solvents and solidify the ink, ensuring strong adhesion to the substrate and optimal electrical conductivity. Curing temperatures and times vary; for example, carbon inks may be dried at 60°C for 30 minutes, while silver inks might require 120°C for 60 minutes [5].
- Insulation and Final Assembly: A final insulating layer is often applied to cover the contact leads, leaving only the active electrode areas and terminal contacts exposed [4]. This step defines the electrochemical active area and protects the conductive tracks.
Table 1: Key Fabrication Steps and Parameters for Screen-Printed Electrodes
| Fabrication Step | Key Parameters | Common Materials/Examples | Impact on Final Product |
|---|---|---|---|
| Substrate Selection | Flexibility, chemical inertness, surface energy | PVC, polyester, polycarbonate, ceramic [4] | Determines mechanical robustness and application suitability (e.g., rigid vs. flexible) |
| Ink Formulation | Conductive material, binder ratio, solvent, viscosity | Graphite, graphene, carbon nanotubes, Ag/AgCl paste [6] [4] | Defines electrical conductivity, electrochemical window, and modifiability |
| Printing Process | Mesh size, squeegee pressure, printing speed | Manual or automated screen-printing systems [5] | Controls pattern resolution, thickness of deposited layer, and manufacturing throughput |
| Curing/Drying | Temperature, time, atmosphere | Oven drying: 60°C for carbon, 120°C for silver [5] | Ensures ink adhesion, solvent removal, and final electrical properties |
Advantages for Decentralized Analysis
SPEs offer a compelling set of advantages that make them ideally suited for decentralized analytical applications, such as on-site pesticide detection [1] [7].
- Portability and Miniaturization: The small size and lightweight nature of SPEs allow them to be integrated into handheld, portable potentiostats, enabling field-deployable analysis outside of traditional laboratories [8] [2].
- Low Cost and Disposability: The mass-production capability of screen printing dramatically reduces the per-unit cost of electrodes. This disposability is critical for avoiding cross-contamination between samples, which is a significant concern in pesticide analysis of complex agricultural or food matrices [1] [4].
- Ease of Use and Mass Production: SPEs are pre-configured, user-friendly devices that require no polishing or pre-treatment before use, simplifying the analytical workflow. The screen-printing technique supports the high-volume, reproducible manufacturing of sensors with consistent performance [1] [8].
- Simplified System Integration: The three-electrode system on a single chip simplifies connection to potentiostats and is readily integrated with fluidic systems for automated analysis, as demonstrated in systems for heavy metal detection [7].
Essential Research Reagents and Materials
The functionality of SPEs, particularly for specialized applications like pesticide sensing, is often enhanced through surface modifications. The following table catalogizes key reagents and materials used in the fabrication and modification of SPEs.
Table 2: Research Reagent Solutions for SPE Fabrication and Modification
| Material Category | Specific Examples | Function in SPE Development |
|---|---|---|
| Conductive Inks | Graphite ink, Silver/Silver Chloride (Ag/AgCl) ink, Carbon nanotube (CNT) ink, Graphene ink [5] [4] | Forms the conductive pathways for the working, counter, and reference electrodes; foundation for electron transfer. |
| Nanomaterials | Gold Nanoparticles (AuNPs), Graphene & its derivatives, Carbon Nanotubes (CNTs), Metal Oxides (e.g., CuO) [9] [4] [3] | Enhances electrochemical sensitivity and surface area; can provide catalytic activity or serve as an immobilization platform. |
| Biorecognition Elements | Acetylcholinesterase (AChE) enzyme, Antibodies, Aptamers, Molecularly Imprinted Polymers (MIPs) [1] [3] | Provides high selectivity for the target analyte (e.g., pesticides) by leveraging specific biological or biomimetic interactions. |
| Polymers & Binders | Chitosan, Polyvinyl alcohol (PVA), Nafion, Polyethylene oxide (PEO) [6] [5] | Used as substrates, hydrogel matrices for entrapment, or binders to improve adhesion and stability of the modified layer. |
| Chemical Modifiers | Prussian Blue, Meldola's Blue, Poly(3,4-ethylenedioxythiophene) polystyrene sulfonate (PEDOT:PSS) [6] [2] | Acts as electrocatalysts or electron mediators to lower working potentials and improve signal-to-noise ratio. |
Experimental Protocol: Fabrication and Modification of SPEs for Pesticide Detection
This protocol provides a detailed methodology for the in-house fabrication of carbon-based SPEs and their subsequent modification with a nanocomposite for the electrochemical detection of organophosphorus pesticides. The workflow is summarized in the diagram below.
Materials and Equipment
- Materials: Polyvinyl chloride (PVC) sheet as substrate; graphite conductive ink (e.g., SC-1010 from ITK); silver/silver chloride (Ag/AgCl) ink (e.g., NT-6307-2 from PERM TOP); chitosan (medium molecular weight); graphene nanoplatelets; acetylcholinesterase (AChE) enzyme; acetylthiocholine (ATCh) iodide; phosphate buffer saline (PBS, 0.1 M, pH 7.4) [5] [4] [3].
- Equipment: Screen-printing apparatus (e.g., Model NSP-1A, YULISHIH INDUSTRIAL Co., Ltd.); precision oven; laboratory potentiostat/ galvanostat; ultrasonic bath.
Step-by-Step Procedure
Part A: Fabrication of Carbon Screen-Printed Electrodes
- Substrate Cleaning: Clean the PVC substrate sheet with dichloromethane (DCM) or ethanol to remove any organic contaminants and ensure good ink adhesion. Allow to dry completely [4].
- Screen Alignment: Secure the patterned screen (designed with a three-electrode layout) over the clean PVC substrate.
- Printing Working and Counter Electrodes: Apply graphite ink to the screen and use a squeegee to spread it evenly, forcing ink through the mesh onto the substrate to form the working and counter electrodes. Repeat this step to ensure a smooth, uniform layer [4].
- Intermediate Curing: Transfer the printed substrate to an oven and cure at 60°C for 30 minutes to dry the carbon ink [5].
- Printing Reference Electrode: Using a clean screen patterned for the reference electrode, apply Ag/AgCl ink to form the reference electrode.
- Final Curing: Cure the complete SPE at a higher temperature of 120°C for 60 minutes to ensure all inks are fully dried and adhered [5].
Part B: Surface Modification with Nanocomposite for Pesticide Sensing
- Nanocomposite Ink Preparation: Prepare a composite ink by dispersing 2 mg/mL of graphene nanoplatelets in a 0.5% chitosan solution (dissolved in 0.1 M acetic acid). Sonicate for 60 minutes to achieve a homogeneous suspension [6].
- Enzyme Solution Preparation: Prepare a solution of AChE enzyme (1 U/μL) in chilled 0.1 M PBS, pH 7.4.
- Electrode Modification: Deposit 5 μL of the graphene-chitosan nanocomposite ink onto the surface of the carbon working electrode and allow it to dry at room temperature. Subsequently, deposit 3 μL of the AChE enzyme solution onto the modified surface and allow it to immobilize under refrigeration (4°C) for 12 hours [3].
Electrochemical Characterization and Validation
- Cyclic Voltammetry (CV): Characterize the modified SPE by performing CV in a 5 mM solution of potassium ferricyanide in 0.1 M KCl. Scan between -0.2 V and +0.6 V at a scan rate of 50 mV/s. A well-modified electrode should show a well-defined, reversible redox peak with an increase in peak current compared to an unmodified electrode, indicating enhanced surface area and electron transfer [4].
- Electrochemical Impedance Spectroscopy (EIS): Further characterize the electrode in the same redox probe solution. Apply a DC potential at the formal potential of the redox couple with a 10 mV AC perturbation across a frequency range of 0.1 Hz to 100 kHz. A significant decrease in the charge transfer resistance (Rₑₜ) observed in the Nyquist plot confirms improved electron transfer kinetics due to the nanocomposite modification [4].
Screen-printed electrodes provide a robust, versatile, and economically viable platform for decentralized analytical sensing. Their design, which integrates all necessary electrodes on a single chip, combined with a scalable fabrication process, makes them indispensable for modern applications ranging from clinical diagnostics to environmental monitoring. As the demand for on-site analysis grows, particularly in fields like pesticide residue monitoring, the role of SPEs is set to expand. Future advancements will likely focus on the development of novel nanomaterial composites and biorecognition elements to further enhance the sensitivity, selectivity, and stability of these devices, solidifying their position as a cornerstone of decentralized analytical science.
The accurate and sensitive detection of pesticide residues is a critical challenge in ensuring food safety, environmental health, and public safety. Traditional analytical methods, while effective, often require sophisticated laboratory equipment, trained personnel, and are time-consuming, limiting their use for rapid, on-site screening [10] [11]. Within the context of a broader thesis on nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, this document establishes the foundational role of specific nanomaterial classes.
Electrochemical biosensors have emerged as predominant tools, offering rapid, sensitive, and cost-effective analysis [12]. The core of these sensors is the transducer, with SPEs being particularly advantageous due to their low cost, portability, ease of mass production, and minimal sample volume requirements [10] [13] [14]. However, the performance of SPEs is substantially enhanced through strategic surface modification with nanomaterials [11]. The integration of noble metals, carbon nanostructures, and metal oxides confers unique physicochemical properties—such as high electrical conductivity, large surface area, and superior catalytic activity—that collectively increase sensor sensitivity, selectivity, and stability [12] [15] [11]. This application note provides a detailed overview of these critical nanomaterials, their properties, and standardized protocols for their application in advanced electrochemical sensing platforms for pesticide detection.
Critical Nanomaterial Classes: Properties and Functions
The enhancement of electrochemical sensors relies on the synergistic properties of various nanomaterials. The table below summarizes the key characteristics and primary functions of the three critical nanomaterial classes in pesticide sensor applications.
Table 1: Critical Nanomaterial Classes for Electrode Modification in Pesticide Sensing
| Nanomaterial Class | Key Properties | Primary Functions in Pesticide Sensing | Common Examples |
|---|---|---|---|
| Noble Metals | High electrical conductivity, excellent catalytic activity, biocompatibility, surface plasmon resonance [12] [11]. | Signal amplification, electrocatalysis of redox reactions, facilitation of electron transfer, label for biorecognition elements [12] [10]. | Gold (Au), Silver (Ag), Platinum (Pt), Palladium (Pd) nanoparticles; Au-Pd bimetallic nanoparticles [10] [16]. |
| Carbon Nanostructures | High surface area, excellent electrical conductivity, mechanical strength, chemical stability, good biocompatibility [17] [12]. | Providing a high-surface-area scaffold, enhancing electron transfer kinetics, increasing biomolecule adsorption, serving as a conductive support [12] [11]. | Graphene, Carbon Nanotubes (CNTs), Carbon Nanofibers (CNFs), Reduced Graphene Oxide (rGO) [17] [12] [10]. |
| Metal Oxides | Catalytic activity, high surface area, tunable electronic properties, semiconducting nature, photocatalytic properties [12]. | Electrocatalysis, signal enhancement for specific analytes, improving sensor stability and selectivity [12]. | Iron Oxide (Fe₃O₄), Titanium Oxide (TiO₂), Zinc Oxide (ZnO) [12]. |
The synergistic combination of these materials in hybrid nanocomposites often yields superior performance. For instance, the hybridization of carbon nanotubes with metal oxide nanoparticles can significantly enhance electron transfer kinetics and sensor sensitivity [12]. Similarly, combining graphene with metal nanoparticles provides a highly conductive and catalytically active platform ideal for immobilizing biological recognition elements [12].
Experimental Protocols for Electrode Modification and Characterization
Standardized Protocol for Nanomaterial Modification of SPEs
The following protocol outlines a generalized procedure for modifying screen-printed electrodes (SPEs) with nanomaterials, which can be adapted for specific material types.
Table 2: The Scientist's Toolkit: Key Reagents and Materials for Electrode Modification
| Item Name | Function/Description | Example Use Case |
|---|---|---|
| Screen-Printed Electrode (SPE) | A miniaturized, disposable electrochemical cell; serves as the foundational transducer [10] [13]. | The base platform for all modifications, typically with carbon, gold, or silver working electrodes. |
| Nanomaterial Suspension | A stable, homogeneous dispersion of the selected nanomaterial in a suitable solvent (e.g., water, ethanol). | The active modifier used in drop-casting to enhance the electrochemical properties of the SPE surface. |
| Phosphate Buffered Saline (PBS) | A buffer solution used to maintain a stable pH during electrochemical measurements and biomolecule immobilization. | Provides a consistent chemical environment for reliable and reproducible electrochemical analysis. |
| Biopolymer (e.g., Chitosan, Nafion) | A polymeric matrix used to entrap and stabilize nanomaterials and biomolecules on the electrode surface [13]. | Acts as a binder and stabilizing agent, preventing nanomaterial leaching and improving film adhesion. |
| Electrochemical Workstation | Instrument for applying controlled potentials and measuring resulting currents for sensor characterization [18]. | Used for Cyclic Voltammetry (CV), Electrochemical Impedance Spectroscopy (EIS), and analytical measurements. |
Procedure:
- SPE Pretreatment: Clean the working electrode surface of the SPE by cycling the potential in a suitable electrolyte (e.g., 0.1 M H₂SO₄ or PBS) using Cyclic Voltammetry (CV) until a stable voltammogram is obtained. This removes any contaminants.
- Suspension Preparation: Disperse the desired nanomaterial (e.g., 1-5 mg) in a solvent (e.g., deionized water, ethanol) to a final volume of 1-10 mL. Sonicate the mixture for 30-60 minutes to achieve a homogeneous suspension [16].
- Modification via Drop-Casting:
- Pipette a precise volume (typically 2-10 µL) of the nanomaterial suspension directly onto the pre-cleaned working electrode surface.
- Allow the electrode to dry under ambient conditions or under a gentle infrared lamp [16]. For more uniform films, controlled drying under a nitrogen stream is recommended to mitigate the "coffee-ring" effect [15].
- Stabilization (Optional): To enhance the stability of the modification layer, apply a thin layer of a biopolymer (e.g., 2-5 µL of 0.5% Nafion or Chitosan solution) over the dried nanomaterial film and allow it to dry.
Alternative Methods: For higher precision and uniformity, other deposition techniques can be employed:
- Electrodeposition: The nanomaterial or metal nanoparticles are deposited onto the SPE surface by applying a constant potential or cycling the potential in a solution containing metal ion precursors (e.g., HAuCl₄, PdCl₂) [15] [16].
- Spin Coating: A small volume of suspension is placed on the electrode, which is then spun at high speed to create a thin, uniform film [15].
- Spray Coating: The suspension is sprayed onto the electrode surface using a carrier gas, allowing for the coating of larger areas [15].
Protocol for Electrochemical Characterization of Modified Electrodes
Characterizing the modified electrode is crucial to confirm successful modification and assess its electrochemical performance.
Materials:
- Potentiostat/Galvanostat (Electrochemical Workstation)
- Modified and unmodified (control) SPEs
- Redox probe solution (e.g., 5 mM [Fe(CN)₆]³⁻/⁴⁻ in 0.1 M KCl)
- Phosphate Buffer Saline (PBS, 0.1 M, pH 7.4)
Procedure:
- Cyclic Voltammetry (CV) in a Redox Probe:
- Immerse the modified SPE in a solution of 5 mM [Fe(CN)₆]³⁻/⁴⁻ in 0.1 M KCl.
- Record a CV scan within a potential window of -0.2 V to 0.6 V (vs. the on-chip reference) at a scan rate of 50 mV/s.
- A successful modification is indicated by an increase in the peak current and a decrease in the peak-to-peak separation (ΔEp) compared to the bare SPE, signifying enhanced electron transfer kinetics [10].
- Electrochemical Impedance Spectroscopy (EIS):
- Using the same redox probe solution, perform EIS at a DC potential corresponding to the formal potential of the redox couple, with a small AC voltage amplitude (e.g., 5-10 mV) over a frequency range from 100 kHz to 0.1 Hz.
- The diameter of the semicircle in the Nyquist plot represents the charge transfer resistance (Rₑₜ). A decrease in Rₑₜ after modification confirms improved conductivity and electron transfer efficiency [10] [16].
The following workflow diagram summarizes the key stages from electrode modification to sensor characterization.
Diagram 1: Workflow for electrode modification and characterization. The process begins with electrode pretreatment, proceeds through one of several modification paths, and concludes with electrochemical validation.
Analytical Performance and Application in Pesticide Detection
The true value of nanomaterial-modified SPEs is demonstrated through their analytical performance in detecting specific pesticides. The following table compiles representative data from the literature showcasing the efficacy of different nanomaterial composites.
Table 3: Analytical Performance of Nanomaterial-Modified Electrodes for Pesticide Detection
| Target Pesticide | Electrode Modification | Detection Technique | Linear Range | Limit of Detection (LOD) | Application |
|---|---|---|---|---|---|
| Organophosphorus (e.g., Paraoxon) | Acetylcholinesterase (AChE) enzyme immobilized on CNT/Nafion-SPE [10] | Amperometry (Enzymatic Inhibition) | Not Specified | Low µM range | Water and Food Samples [10] |
| Organophosphorus & Carbamate | Multi-enzyme system (AChE, BChE, Tyrosinase) on SPE [10] | Amperometry (Enzymatic Inhibition) | Not Specified | Not Specified | Multi-analyte Screening [10] |
| Fenobucarb | Graphene Nanoribbons-Ionic Liquid-Cobalt Phthalocyanine/SPE [11] | Flow Injection Analysis | Not Specified | High Sensitivity Reported | Not Specified [11] |
| Diclofenac Sodium (Model Drug) | Au-Pd Bimetallic NPs/Halloysite/PGE [16] | Differential Pulse Voltammetry (DPV) | 1 – 100 µM | 0.047 µM | Proof of Concept for Sensor Design [16] |
The primary sensing mechanisms for pesticides include:
- Enzymatic Inhibition: The most common route, where pesticides like organophosphates and carbamates inhibit enzymes such as acetylcholinesterase (AChE). The decrease in enzymatic activity, measured electrochemically, is proportional to the pesticide concentration [10].
- Direct Electrochemical Detection: Electroactive pesticides can be directly oxidized or reduced on the catalytically active surface of the modified electrode, with the current response being proportional to concentration [10] [11].
The integration of noble metals, carbon nanostructures, and metal oxides into screen-printed electrodes represents a powerful strategy for advancing electrochemical sensor technology for pesticide analysis. The protocols and data summarized in this application note provide a framework for researchers to fabricate and characterize high-performance, nanomaterial-modified sensing platforms. The demonstrated enhancements in sensitivity, selectivity, and portability make these devices compelling tools for on-site monitoring, addressing critical needs in food safety and environmental protection.
Future developments in this field are likely to focus on several key areas:
- Advanced Nanocomposites: Designing more sophisticated multi-functional nanocomposites that leverage synergistic effects for even greater analytical performance [12] [11].
- Integration with Smart Technology: Coupling these sensors with smartphone-based readouts and data transmission for real-time, geographically tagged analysis [11].
- Lab-on-a-Chip (LOC) Systems: Incorporating modified SPEs into fully integrated microfluidic LOC devices that automate sample preparation and analysis, enhancing usability and reliability for field deployment [11].
- Machine Learning (ML): Employing ML algorithms to analyze complex electrochemical data, predict sensor performance based on fabrication parameters, and improve the accuracy of multi-analyte detection [18].
By continuing to refine these materials and methodologies, the scientific community can develop next-generation analytical devices that are not only highly effective but also accessible and practical for widespread use.
The escalating need for global food production has led to the extensive use of pesticides, including organophosphates (OPs), carbamates, and neonicotinoids [19]. While effective for crop protection, their persistence in the environment and subsequent contamination of food and water sources pose significant health risks to humans, ranging from neurological disorders to carcinogenic effects [10] [20]. This has spurred the development of rapid, sensitive, and cost-effective detection methods, moving beyond traditional techniques like chromatography and mass spectrometry [10].
Electrochemical sensing, particularly using nanomaterial-modified screen-printed electrodes (SPEs), has emerged as a powerful analytical tool for pesticide monitoring [1] [10]. These sensors leverage the distinct electrochemical properties of different pesticide classes, enabling the design of highly specific and sensitive detection platforms. This application note details the electrochemical behaviors of major pesticide classes and provides standardized protocols for their analysis using modified SPEs, serving as a practical guide for researchers developing advanced electrochemical sensors.
Electrochemical Properties and Detection Mechanisms by Pesticide Class
The detection strategy for a pesticide is fundamentally guided by its molecular structure and intrinsic electrochemical properties. The following sections delineate the characteristics and sensing mechanisms for the three primary insecticide classes.
Organophosphates (OPs)
Mechanism of Toxicity: OPs irreversibly inhibit the enzyme acetylcholinesterase (AChE), leading to the accumulation of the neurotransmitter acetylcholine and resulting in neurotoxicity [21] [22].
Detection Mechanisms:
- Enzyme Inhibition-Based Sensing: This is the most common strategy for OP detection. The principle involves immobilizing AChE on the electrode surface. The active enzyme hydrolyzes its substrate, acetylthiocholine (ATCh), producing an electroactive product, thiocholine, which generates a measurable current. Upon exposure to OPs, the enzyme is inhibited, leading to a reduction in the catalytic current that is proportional to the OP concentration [10] [23].
- Enzymatic Catalysis-Based Sensing: Certain OPs can be directly hydrolyzed by enzymes like organophosphate hydrolase (OPH). The hydrolysis of OPs like paraoxon and parathion produces p-nitrophenol (p-NP), an electroactive species that can be detected amperometrically [21] [22].
Table 1: Electrochemical Detection of Select Organophosphates.
| Pesticide | Detection Mechanism | Electrode Modification | Linear Range | Limit of Detection (LOD) |
|---|---|---|---|---|
| Chlorpyrifos | AChE Inhibition | CuNWs/rGO on SPCE [23] | 10 - 200 µg/L | 3.14 µg/L |
| Paraoxon | OPH Catalysis | Engineered Microbial System [22] | Sub-µM range | Not Specified |
| Methyl Parathion | OPH Catalysis | MPH/Silica-Gold-CNT Composite [21] | Refer to [21] | Refer to [21] |
Carbamates
Mechanism of Toxicity: Similar to OPs, carbamates are AChE inhibitors, but their action is reversible, which generally makes them less toxic to mammals than OPs [20] [24].
Detection Mechanisms:
- Enzyme Inhibition-Based Sensing: Analogous to OPs, carbamates can be detected by measuring their inhibitory effect on the activity of AChE or other enzymes like tyrosinase immobilized on the electrode [10].
- Direct Electrochemical Detection: Some carbamates, or their hydrolysis products, are electroactive. For instance, the carbamate carbaryl can be directly oxidized on suitably modified electrodes, allowing for direct quantification without an enzyme [20].
Table 2: Electrochemical Detection of Select Carbamates.
| Pesticide | Detection Mechanism | Electrode Modification | Linear Range | Limit of Detection (LOD) |
|---|---|---|---|---|
| Carbofuran | AChE Inhibition | AuNPs/MWCNT on SPCE [20] | 0.5 - 100 µM | 0.21 µM |
| Carbaryl | Direct Oxidation | Molecularly Imprinted Polymer (MIP) [20] | 1 - 100 µM | 0.8 µM |
| Aldicarb, Propoxur | Voltammetric Analysis | Glassy Carbon Electrode (GCE) [20] | Varies by compound | Varies by compound |
Neonicotinoids
Mechanism of Toxicity: Neonicotinoids act as agonists on the nicotinic acetylcholine receptors (nAChR) in the central nervous system of insects, causing overstimulation and death. They exhibit selective toxicity towards insects over mammals due to higher affinity for insect nAChRs [19] [24].
Detection Mechanisms:
- Direct Electrochemical Detection: This is the predominant strategy for neonicotinoids. Many, such as imidacloprid and acetamiprid, contain electroactive functional groups (e.g., nitro groups) that can be directly reduced or oxidized on the electrode surface. The resulting current is directly proportional to concentration [19] [24].
- Aptasensors and Immunosensors: Specific aptamers or antibodies are used as recognition elements for neonicotinoids. The binding event is then transduced into an electrochemical signal, often using EIS, providing high specificity [24].
Table 3: Electrochemical Detection of Select Neonicotinoids.
| Pesticide | Detection Mechanism | Electrode Modification | Linear Range | Limit of Detection (LOD) |
|---|---|---|---|---|
| Imidacloprid | Direct Reduction | Fe-rich FeCoNi-MOF [24] | 0.005 - 20 µM | 0.26 nM |
| Acetamiprid | Aptasensor | rGO/β-cyclodextrin polymer [24] | 1 pM - 1 µM | 0.34 pM |
| Thiamethoxam | Direct Detection | Boron-Doped Diamond [24] | 0.49 - 7.36 µM | 0.12 µM |
Experimental Protocols
Protocol 1: Detection of Organophosphates via an AChE Inhibition Biosensor
This protocol details the construction of a biosensor for chlorpyrifos detection using an AChE enzyme inhibition approach on a CuNWs/rGO-modified SPCE [23].
The Scientist's Toolkit: Key Research Reagents
| Reagent / Material | Function in the Experiment |
|---|---|
| Screen-Printed Carbon Electrode (SPCE) | Disposable, portable electrochemical transducer platform. |
| Reduced Graphene Oxide (rGO) | Enhances electrical conductivity and provides a large surface area for biomolecule immobilization. |
| Copper Nanowires (CuNWs) | Improves electrocatalytic activity and electron transfer, particularly for thiocholine oxidation. |
| Acetylcholinesterase (AChE) | Biological recognition element; its inhibition is measured. |
| Acetylthiocholine (ATCh) | Enzyme substrate; hydrolysis produces electroactive thiocholine. |
| Phosphate Buffer Saline (PBS) | Provides a stable pH environment for the enzymatic reaction. |
| Glutaraldehyde | Crosslinking agent for enzyme immobilization on the electrode surface. |
Procedure:
- Electrode Modification:
- Prepare a homogeneous dispersion of CuNWs/rGO nanocomposite in a suitable solvent (e.g., DMF).
- Drop-cast a precise volume (e.g., 5 µL) of the nanocomposite dispersion onto the working electrode area of the SPCE.
- Allow the solvent to evaporate completely at room temperature to form a stable modified film.
- Enzyme Immobilization:
- Prepare a solution of AChE (e.g., 0.5 U/µL) in a mild phosphate buffer (pH 7.4).
- Apply the AChE solution onto the modified SPCE surface.
- Use glutaraldehyde vapor to cross-link and stabilize the enzyme layer.
- Rinse the electrode gently with PBS to remove any unbound enzyme.
- Electrochemical Measurement (Baseline):
- Place the modified SPCE in an electrochemical cell containing PBS (pH 7.4) and ATCh.
- Record a cyclic voltammogram (CV) or a chronoamperogram. A clear oxidation peak or steady-state current corresponding to the enzymatic production of thiocholine should be observed. This is the baseline signal (I₀).
- Inhibition (Pesticide Detection):
- Incubate the biosensor in a sample solution containing the target organophosphate (e.g., chlorpyrifos) for a fixed time (e.g., 10-15 minutes).
- Gently rinse the electrode with PBS to remove the pesticide solution.
- Electrochemical Measurement (Post-Inhibition):
- Record the CV or chronoamperogram again under the same conditions as in Step 3. The measured current (I) will be lower due to enzyme inhibition.
- Quantification:
- The percentage of enzyme inhibition is calculated as: Inhibition (%) = [(I₀ - I) / I₀] × 100.
- The inhibition percentage is proportional to the logarithm of the pesticide concentration and can be plotted to create a calibration curve.
The following workflow illustrates the key steps in this biosensing protocol:
Diagram 1: AChE Inhibition Biosensor Workflow.
Protocol 2: Direct Electrochemical Detection of a Neonicotinoid
This protocol outlines the direct voltammetric detection of imidacloprid, leveraging the electrochemical reduction of its nitro group on a nanomaterial-modified electrode [19] [24].
Procedure:
- Electrode Modification:
- Modify the SPCE or GCE with the selected nanomaterial (e.g., Fe-rich FeCoNi-MOF, graphene oxide, or boron-doped diamond).
- The modification can be achieved via drop-casting or electrodeposition to form a uniform catalytic layer.
- Electrochemical Measurement:
- Prepare standard solutions of imidacloprid in a supporting electrolyte (e.g., Britton-Robinson buffer, pH 7.0).
- Transfer the analyte solution to the electrochemical cell.
- Using the modified working electrode, apply a square-wave voltammetry (SWV) or differential pulse voltammetry (DPV) potential sweep in a negative direction (e.g., from -0.4 V to -1.0 V).
- The reduction of the nitro group (-NO₂) to the hydroxylamine group (-NHOH) will produce a characteristic cathodic peak.
- Calibration and Quantification:
- Record voltammograms for a series of standard solutions with increasing imidacloprid concentrations.
- Plot the peak current intensity against the concentration to generate a calibration curve for quantitative analysis of unknown samples.
The logical relationship between the analyte's structure and the detection signal is as follows:
Diagram 2: Direct Detection Signaling Logic.
The distinct electrochemical properties of different pesticide classes dictate the design and application of effective sensing strategies. Organophosphates and carbamates are predominantly detected via enzyme inhibition pathways, while neonicotinoids are often quantified through direct electron transfer involving their nitro group. The use of nanomaterial-modified SPEs is a cornerstone of modern electrochemical pesticide analysis, providing enhanced sensitivity, selectivity, and portability. The protocols outlined herein offer a foundational framework for researchers to develop and optimize robust electrochemical sensors for environmental monitoring and food safety assurance. Future perspectives point towards the increased integration of novel biorecognition elements like aptamers, the development of multi-analyte arrays, and the creation of fully integrated, field-deployable devices.
The core of any advanced electrochemical (bio)sensor is its recognition element, a biological or biomimetic molecule designed to interact specifically with a target analyte. The selectivity and sensitivity of the sensor are fundamentally determined by the affinity and properties of this element. For the analysis of pesticides using nanomaterial-modified screen-printed electrodes (SPEs), four primary classes of recognition elements are predominantly employed: enzymes, antibodies, aptamers, and Molecularly Imprinted Polymers (MIPs). Screen-printed electrodes serve as an ideal platform for such sensing due to their cost-effectiveness, portability for on-site analysis, and ease of modification with various nanomaterials and recognition elements [25] [13]. The integration of nanomaterials like gold nanoparticles, carbon nanotubes, and graphene oxide further enhances the electrochemical performance by improving electron transfer, increasing surface area, and providing a scaffold for the immobilization of these recognition elements [26] [27] [28]. This document provides detailed application notes and experimental protocols for the integration of these four key recognition elements within the context of a thesis focused on nanomaterial-modified SPEs for pesticide analysis.
The choice of recognition element dictates the design, performance, and application range of the sensor. The following table offers a structured comparison of these elements to guide selection.
Table 1: Comparative Analysis of Recognition Elements for Pesticide Sensing on SPEs
| Recognition Element | Mechanism of Action | Key Advantages | Inherent Limitations | Typical Electrochemical Technique |
|---|---|---|---|---|
| Enzymes | Catalytic transformation or inhibition of the target pesticide. | High catalytic activity; Well-established protocols; Reusable sensors. | Susceptible to environmental conditions (pH, T); Limited enzyme stability; Broad specificity for inhibitor classes. | Amperometry, Chronoamperometry [25] |
| Antibodies | Specific immunochemical binding (antigen-antibody). | Exceptional specificity and affinity; Wide variety commercially available. | Animal-derived production; Batch-to-batch variability; Sensitive to denaturation; Irreversible binding. | Electrochemical Impedance Spectroscopy (EIS) [25] [26] |
| Aptamers | Conformational change upon binding to a specific target. | Synthetic production (low cost, high stability); Reversible binding; Modifiable chemistry. | In vitro selection process (SELEX) can be complex; Susceptibility to nuclease degradation in biofluids. | EIS, Differential Pulse Voltammetry (DPV), Square Wave Voltammetry (SWV) [27] [29] |
| Molecularly Imprinted Polymers (MIPs) | Selective rebinding to synthetic, template-shaped cavities. | High physical/chemical robustness; Applicable to a wide range of targets; Long shelf-life. | Risk of incomplete template removal; Heterogeneous binding sites; Optimization can be complex. | DPV, Cyclic Voltammetry (CV), EIS [30] [31] |
Detailed Application Notes & Experimental Protocols
Enzymatic Sensors: Acetylcholinesterase-based Inhibition Assay
Principle: This protocol is based on the inhibition of the enzyme acetylcholinesterase (AChE) by organophosphorus and carbamate pesticides. The active enzyme hydrolyzes its substrate, producing an electroactive product. The presence of the pesticide inhibits AChE, leading to a measurable decrease in the electrochemical signal, which is proportional to the pesticide concentration [25].
Experimental Protocol:
SPE Nanomaterial Modification:
- Clean the bare carbon SPE by performing 10 cycles of Cyclic Voltammetry (CV) in 0.1 M H₂SO₄ from 0 V to +1.2 V (vs. Ag/AgCl reference).
- Deposit 5 µL of a graphene oxide (GO) dispersion (1 mg/mL) onto the working electrode and allow it to dry at room temperature.
- Electrochemically reduce the GO to rGO by performing CV in 0.1 M phosphate buffer saline (PBS), pH 7.4, for 15 scans.
Enzyme Immobilization:
- Prepare an immobilization mixture containing 5 µL of AChE (2 U/µL), 5 µL of bovine serum albumin (BSA, 1% w/v), and 2 µL of glutaraldehyde (0.25% v/v).
- Deposit 8 µL of this mixture onto the rGO/SPE and incubate at 4°C for 1 hour.
- Rinse the modified electrode thoroughly with PBS (pH 7.4) to remove any unbound enzyme.
Pesticide Incubation (Inhibition Step):
- Immerse the AChE/rGO/SPE in a solution containing the target pesticide (e.g., chlorpyrifos) for 15 minutes.
- Rinse gently with PBS.
Electrochemical Measurement:
- Transfer the sensor to an electrochemical cell containing 10 mL of PBS (pH 7.4) with 1.0 mM acetylthiocholine (ATCh), the enzyme substrate.
- Perform Chronoamperometry at an applied potential of +0.5 V for 60 seconds.
- The current generated from the oxidation of the enzymatic product (thiocholine) is recorded. The percentage of inhibition is calculated as:
% Inhibition = [(I₀ - I)/I₀] × 100, where I₀ and I are the currents before and after incubation with the pesticide, respectively.
Diagram 1: AChE Inhibition Assay Workflow
Immunosensors: Antibody-based Detection with Nanomaterial Labels
Principle: This protocol describes a sandwich-type electrochemical immunosensor. A capture antibody is immobilized on the SPE. The target pesticide (acting as an antigen) is bound, and is subsequently recognized by a second detection antibody conjugated to a nanomaterial label, such as gold nanoparticles (AuNPs). The electrochemical signal from the AuNP label is quantified via anodic stripping voltammetry, providing high sensitivity [26].
Experimental Protocol:
SPE Functionalization and Capture Antibody Immobilization:
- Modify the SPE working electrode with a dispersion of multi-walled carbon nanotubes (MWCNTs) to enhance surface area and conductivity.
- Activate the surface by applying a potential of +1.7 V in 0.1 M NaOH for 60 seconds.
- Deposit 10 µL of a solution containing the capture antibody (e.g., anti-atrazine IgG, 10 µg/mL in PBS, pH 7.4) and incubate for 12 hours at 4°C.
- Block non-specific binding sites by applying 10 µL of BSA (1% w/v) for 1 hour at room temperature. Rinse with PBS.
Immunoassay Procedure:
- Incubate the modified SPE with 50 µL of the sample containing the target pesticide for 30 minutes at 37°C. Rinse.
- Incubate the sensor with 50 µL of the AuNP-labeled detection antibody solution for 30 minutes at 37°C. Rinse thoroughly.
Electrochemical Detection:
- Place the immunosensor in an electrochemical cell containing 0.1 M HCl.
- Apply a pre-concentration potential of +1.0 V for 120 seconds to oxidize and dissolve the AuNPs into AuCl₄⁻ ions.
- Perform Anodic Stripping Voltammetry (ASV) by scanning from +0.2 V to +1.0 V.
- The sharp oxidation peak current at ~+0.65 V is directly proportional to the concentration of Au³⁺ ions, which in turn is proportional to the concentration of the captured pesticide.
Aptasensors: Label-free Impedimetric Detection
Principle: This protocol utilizes an aptamer that undergoes a conformational change upon binding to its target pesticide. This change alters the interfacial properties of the electrode surface, which is measured as a change in charge transfer resistance (Rcₜ) using Electrochemical Impedance Spectroscopy (EIS) [27] [29].
Experimental Protocol:
SPE Modification and Aptamer Immobilization:
- Modify the SPE with a nanocomposite of gold nanoparticles and reduced graphene oxide (AuNP-rGO) to create a highly conductive platform with ample sites for thiol-binding.
- Prepare a 1 µM solution of the thiolated aptamer specific to the target (e.g., tetracycline) in Tris-EDTA buffer.
- Deposit 10 µL of the aptamer solution onto the AuNP-rGO/SPE and incubate overnight at 4°C to form a self-assembled monolayer via Au-S bonds.
- Rinse and then block the surface with 6-mercapto-1-hexanol (1 mM) for 1 hour to passivate unbound gold sites.
Target Binding and EIS Measurement:
- Record the EIS spectrum of the aptasensor in a 5 mM [Fe(CN)₆]³⁻/⁴⁻ redox probe solution (in 0.1 M KCl) as a baseline. Parameters: DC potential of +0.22 V, amplitude of 5 mV, frequency range from 0.1 Hz to 100 kHz.
- Incubate the aptasensor with the sample solution containing the pesticide for 20 minutes.
- Rinse the sensor gently and record the EIS spectrum again under identical conditions.
- The increase in the diameter of the semicircle in the Nyquist plot, corresponding to an increase in Rcₜ, is used to quantify the pesticide concentration.
Diagram 2: Aptamer-based EIS Sensing Workflow
Molecularly Imprinted Polymer Sensors
Principle: MIPs are synthetic polymers with cavities complementary in shape, size, and functional groups to the target molecule (the template). This protocol involves the in-situ electropolymerization of a monomer around the template pesticide on the SPE surface. After template removal, the resulting cavities selectively rebind the pesticide from samples [30] [31].
Experimental Protocol:
SPE Pre-treatment and MIP Formation:
- Clean the carbon SPE via CV in 0.5 M H₂SO₄.
- Prepare a polymerization solution containing the template (e.g., 5 mM paraoxon), the functional monomer (e.g., 20 mM o-phenylenediamine), and 0.1 M PBS (pH 7.0).
- Deposit 15 µL of this solution onto the working electrode.
- Perform Electropolymerization by applying CV between -0.2 V and +0.8 V for 15 cycles at a scan rate of 50 mV/s.
Template Removal:
- Carefully wash the polymerized SPE with a mixture of methanol and acetic acid (9:1, v/v) under gentle stirring for 15 minutes to extract the template molecules, leaving behind specific cavities.
- Rinse extensively with PBS until a stable background CV signal is obtained.
Rebinding and Detection:
- Incubate the MIP/SPE in the sample solution for 15 minutes to allow the pesticide to rebind to the cavities.
- Rinse the sensor to remove non-specifically bound molecules.
- Transfer the sensor to a clean electrochemical cell containing a redox probe (e.g., 5 mM [Fe(CN)₆]³⁻/⁴⁻).
- Perform DPV. The binding of the non-electroactive pesticide into the MIP cavities hinders the diffusion of the redox probe to the electrode surface, resulting in a decrease in the peak current, which is correlated to the pesticide concentration.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagents and Materials for Sensor Development
| Item Name | Function / Application | Example Specifications / Notes |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, miniaturized electrochemical cell transducers. | Ceramic or plastic substrates with carbon, gold, or silver ink working electrodes. |
| Acetylcholinesterase (AChE) | Enzyme for inhibition-based detection of OPs and carbamates. | Source: Electric eel; Activity: >1000 U/mg. Store at -20°C. |
| Anti-pesticide Antibodies | Capture and detection elements for immunosensors. | Monoclonal antibodies preferred for specificity. Requires cold chain storage. |
| DNA/RNA Aptamers | Synthetic recognition elements for aptasensors. | Thiol- or amino-modified for surface immobilization. HPLC purified. |
| Gold Nanoparticles (AuNPs) | Nanomaterial for electrode modification and as an electrochemical label. | ~20 nm diameter, functionalized with streptavidin or antibodies. |
| Graphene Oxide / Reduced Graphene Oxide | Nanomaterial to enhance electrode conductivity and surface area. | Aqueous dispersion, 1-5 mg/mL. |
| o-Phenylenediamine | Functional monomer for electropolymerization of MIP films. | Used in molecular imprinting for phenolic or aromatic targets. |
| Electrochemical Redox Probes | Mediators for signal generation in EIS, DPV, and CV. | 5 mM Potassium ferri/ferrocyanide ([Fe(CN)₆]³⁻/⁴⁻) in 0.1 M KCl. |
| Glutaraldehyde | Crosslinking agent for covalent immobilization of proteins. | Typically used as a 0.25-2.5% (v/v) solution. Handle with care. |
| 6-Mercapto-1-hexanol | Backfiller molecule for Au surfaces to minimize non-specific binding. | Used in aptamer-based sensors to create a well-ordered SAM. |
Performance Data and Analysis
The following table summarizes representative performance metrics achievable with different recognition elements on nanomaterial-modified SPEs, as reported in the literature.
Table 3: Exemplary Performance Metrics for Pesticide Detection
| Recognition Element | Target Pesticide | Nanomaterial Used | Detection Limit | Linear Range | Reference Technique |
|---|---|---|---|---|---|
| AChE (Enzyme) | Chlorpyrifos | Reduced Graphene Oxide | 0.5 ng/L | 1-1000 ng/L | Chronoamperometry [25] |
| Anti-atrazine Antibody | Atrazine | Gold Nanoparticles / CNTs | 0.01 µg/L | 0.05–10 µg/L | ASV [26] |
| Tetracycline Aptamer | Tetracycline | AuNP-rGO nanocomposite | 0.1 nM | 1 nM - 1 µM | EIS [28] [29] |
| MIP (o-PDA polymer) | Paraoxon | Prussian Blue / Carbon Black | 0.8 nM | 5 nM - 5 µM | DPV [31] |
The integration of enzymes, antibodies, aptamers, and MIPs with nanomaterial-modified SPEs provides a powerful and versatile toolbox for advanced pesticide analysis. The choice of the optimal recognition element depends on the specific requirements of the analysis, including the target pesticide, required sensitivity and specificity, sample matrix, and intended use (e.g., one-time field testing vs. continuous monitoring). Enzymes offer a well-understood, catalytic approach ideal for class-specific screening. Antibodies provide unparalleled specificity for individual compounds in a sandwich format. Aptamers present a synthetic, stable, and flexible alternative, excellent for label-free and reversible sensing. MIPs deliver extreme robustness and are suitable for harsh environments and a wide range of targets. A key trend in this field is the development of hybrid systems, such as MIP-aptamer composites, which aim to harness the synergistic advantages of multiple recognition elements to create sensors with superior performance, moving laboratory research closer to real-world deployment [30] [31].
The accurate detection of pesticide residues in food and environmental samples represents a critical challenge in analytical chemistry, directly impacting public health and food safety. Traditional methods, such as chromatography, are often constrained by the need for costly equipment, specialized laboratory settings, and lengthy analysis times [3] [32]. Within this context, electrochemical biosensors based on screen-printed electrodes (SPEs) have emerged as a powerful alternative, offering portability, cost-effectiveness, and the potential for rapid, on-site analysis [1] [2]. The integration of nanomaterials into these sensing platforms has been pivotal in overcoming limitations of sensitivity and selectivity, leading to a transformative leap in their analytical performance [32] [33]. This application note details the fundamental mechanisms through which nanomaterials enhance sensor function, provides a validated experimental protocol for electrode modification and pesticide detection, and outlines the essential toolkit for researchers in this field. The content is specifically framed within ongoing thesis research focused on developing advanced nanomaterial-modified SPEs for pesticide analysis.
Core Enhancement Mechanisms of Nanomaterials
Nanomaterials enhance biosensor performance through several interconnected physical and chemical mechanisms. Their unique properties, such as high surface area-to-volume ratio and quantum effects, directly improve the critical parameters of sensor function.
Table 1: Core Enhancement Mechanisms of Nanomaterials in Electrochemical Sensors
| Enhancement Mechanism | Key Nanomaterials Involved | Primary Effect on Sensor Performance |
|---|---|---|
| Increased Electroactive Surface Area | Carbon nanotubes (CNTs), Graphene, Gold Nanoparticles (AuNPs) | Enhances analyte loading and reaction sites, boosting signal intensity and sensitivity [32] [33]. |
| Enhanced Electron Transfer Kinetics | CNTs, Graphene, Metal Nanoparticles | Acts as an electron "bridge" or conduit, facilitating faster electron shuttling between the biorecognition element and the electrode surface [1] [34]. |
| Catalytic Activity | Metal Oxides (e.g., CuO), Nanozymes, Single-Atom Catalysts (SACs) | Lowers oxidation/reduction overpotentials, improves reaction efficiency, and enables signal amplification [3]. |
| Biorecognition Immobilization | AuNPs, CNTs, Nanohybrids | Provides a stable and favorable microenvironment for anchoring enzymes, antibodies, or aptamers, preserving their bioactivity [3] [32]. |
The synergy of these mechanisms is illustrated in the following diagram, which maps the logical pathway from nanomaterial properties to the final sensor performance metrics.
Quantitative Performance of Nanomaterial-Based Sensors
The practical impact of these enhancement mechanisms is reflected in the superior analytical performance of nanomaterial-based sensors. The following table compiles data from a systematic review of recent research, showcasing the low detection limits achieved for various pesticides in food matrices.
Table 2: Analytical Performance of Selected Nanomaterial-Based Biosensors for Pesticide Detection in Food [32]
| Nanomaterial | Biorecognition Element | Pesticide | Limit of Detection (LOD) | Food Matrix |
|---|---|---|---|---|
| Gold Nanoparticles (AuNPs) | Acetylcholinesterase (AChE) | Organophosphorus (class) | 19–77 ng L⁻¹ | Apple, Cabbage |
| Gold Nanoparticles (AuNPs) | AChE | Methomyl | 81 ng L⁻¹ | Apple, Cabbage |
| Gold Nanoparticles (AuNPs) | AChE | Carbamate (class) | 1.0 nM | Fruit |
| Gold Nanoparticles (AuNPs) | Aptamer | Chlorpyrifos | 36 ng L⁻¹ | Apple, Pak choi |
| Gold Nanoparticles (AuNPs) | Antibody | Chlorpyrifos | 0.07 ng L⁻¹ | Chinese cabbage, Lettuce |
| Nanohybrids | Various | Various | < Maximum Residue Limits | Various fruits/vegetables |
Experimental Protocol: AChE-based Sensor for Organophosphorus Pesticides
This protocol provides a detailed methodology for fabricating a nanomaterial-enhanced acetylcholinesterase (AChE) biosensor for the detection of organophosphorus pesticides (OPs) in fruit juice samples, based on established procedures in the literature [35] [3] [32].
Principle
The sensor operates on an enzyme inhibition mechanism. The immobilized AChE enzyme catalyzes the hydrolysis of acetylthiocholine (ATCh), producing thiocholine. Thiocholine is then electrochemically oxidized at the nanomaterial-modified SPE surface, generating a measurable amperometric current. The presence of OPs inhibits AChE activity, leading to a reduction in the generated thiocholine and a consequent decrease in the electrochemical signal, which is proportional to the pesticide concentration.
Reagents and Materials
- Screen-Printed Electrodes (SPEs): Carbon-based three-electrode systems.
- Nanomaterial Dispersion: e.g., multi-walled carbon nanotubes (MWCNTs, 1 mg/mL in DMF) or graphene oxide (GO, 1 mg/mL in water).
- Biorecognition Element: Acetylcholinesterase (AChE) from Electrophorus electricus.
- Enzyme Substrate: Acetylthiocholine chloride (ATCh) or iodide (ATCHI).
- Buffer: Phosphate Buffered Saline (PBS, 0.1 M, pH 7.4).
- Crosslinker: Glutaraldehyde solution (0.25% v/v).
- Standard Solutions: Parathion, chlorpyrifos, or malathion standards for calibration.
- Real Samples: Fruit juice (e.g., apple juice) filtered and diluted 1:1 with PBS.
Procedure
Step 1: Electrode Modification and Nanomaterial Deposition
- Pretreatment: Activate the bare carbon SPE by cycling the potential in a suitable redox couple or by applying a constant potential in acidic solution to generate oxygenated functional groups [34].
- Modification: Pipette 5 µL of the well-dispersed nanomaterial suspension (e.g., MWCNTs) directly onto the working electrode surface.
- Drying: Allow the electrode to dry at room temperature for 45-60 minutes, forming a uniform film. Rinse gently with distilled water to remove loosely bound material.
Step 2: Enzyme Immobilization
- Preparation: Mix 10 µL of AChE solution (5 U/mL) with 10 µL of a 0.25% glutaraldehyde solution.
- Immobilization: Immediately deposit 5 µL of the AChE-glutaraldehyde mixture onto the nanomaterial-modified working electrode.
- Curing: Let the electrode sit for 1 hour at 4°C to allow for complete cross-linking and enzyme immobilization.
- Storage: Store the prepared biosensors at 4°C in PBS when not in use.
Step 3: Electrochemical Measurement and Pesticide Detection
- Baseline Signal: Place the modified AChE-biosensor in an electrochemical cell containing 10 mL of PBS and 1 mM ATCh. Record the steady-state amperometric current (i₀) at a fixed potential (typically +0.7 V vs. Ag/AgCl reference).
- Inhibition: Incubate the biosensor for 10 minutes in a sample solution (standard or real sample) containing the target OP pesticide.
- Post-Inhibition Signal: Rinse the electrode gently with PBS and measure the amperometric current (i₁) again under the same conditions as in Step 3.1.
- Analysis: The percentage of enzyme inhibition is calculated as: % Inhibition = [(i₀ - i₁) / i₀] × 100. The pesticide concentration is determined by interpolating this value into a pre-established calibration curve.
The workflow for this experimental protocol is summarized in the following diagram:
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful development of a nanomaterial-modified SPE biosensor requires a carefully selected set of materials. The following table lists key reagents and their specific functions within the experimental workflow.
Table 3: Essential Research Reagent Solutions for Sensor Fabrication
| Item | Function / Role in the Experiment |
|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, portable, and mass-producible platform integrating working, reference, and counter electrodes [1] [2]. |
| Carbon Nanotubes (CNTs) / Graphene | High-conductivity nanomaterials that provide a large surface area for enzyme loading and facilitate electron transfer, significantly enhancing signal response [32] [34]. |
| Gold Nanoparticles (AuNPs) | Excellent biocompatibility and conductivity; often used to immobilize biomolecules via Au-S bonds and to enhance electrochemical signals [32] [36]. |
| Acetylcholinesterase (AChE) Enzyme | The primary biorecognition element whose activity is inhibited by organophosphorus and carbamate pesticides, forming the basis of the detection mechanism [3] [32]. |
| Acetylthiocholine (ATCh) | Enzyme substrate; its hydrolysis by AChE produces thiocholine, which is electrochemically oxidized to generate the analytical signal [3]. |
| Glutaraldehyde | A crosslinking agent used to create stable covalent bonds between the enzyme (AChE) and the nanomaterial-modified electrode surface [32]. |
| Phosphate Buffered Saline (PBS) | Provides a stable pH and ionic strength environment for maintaining enzyme activity and for all electrochemical measurements [32]. |
Fabrication Techniques and Detection Methodologies for Practical Pesticide Analysis
The functionalization of transducer surfaces is a critical step in the development of highly sensitive and selective electrochemical sensors. Within the context of screen-printed electrode (SPE)-based platforms for pesticide analysis, the method of applying nanomaterials and biorecognition elements directly governs the analytical performance of the resulting biosensor [9] [10]. SPEs provide a versatile and disposable foundation, but their inherent capabilities are substantially enhanced through deliberate modification strategies that increase effective surface area, improve electron transfer kinetics, and allow for the stable immobilization of specific bioreceptors [34] [10].
This protocol details three cornerstone modification techniques—electrodeposition, drop-casting, and chemical immobilization—tailored for the construction of nanomaterial-enhanced biosensors for pesticide detection. These methods facilitate the creation of a sensitive transduction interface and ensure the robust attachment of biological components such as enzymes, antibodies, or aptamers, which are essential for selective target recognition [32] [10]. The strategic integration of nanomaterials like gold nanoparticles (AuNPs), carbon nanotubes (CNTs), and graphene oxide (GO) is emphasized, as they are pivotal in amplifying the electrochemical signal and lowering detection limits to clinically and environmentally relevant concentrations [9] [32].
Theoretical Foundations of Modification Techniques
The choice of modification technique is governed by the desired properties of the nanomaterial film and the nature of the biological element to be immobilized. Each method presents distinct advantages regarding film uniformity, adhesion strength, processing time, and compatibility with sensitive biomolecules.
Electrodeposition leverages electrochemical principles to precisely control the nucleation and growth of a material onto the electrode surface from a precursor solution. Applying a controlled potential or current density allows for the controlled reduction of metal ions (e.g., Au³⁺, Ag⁺) to form a nanostructured layer. This method typically yields films with strong adhesion and excellent electrical connectivity to the electrode surface, which is crucial for efficient electron transfer [37]. The morphology, particle size, and density of the deposited nanomaterial can be finely tuned by varying key parameters such as the applied potential, deposition time, and the composition of the electrolyte solution [10].
Drop-Casting is a straightforward physical adsorption technique where a small, defined volume of nanomaterial dispersion is pipetted directly onto the working electrode surface and allowed to dry. Its primary advantages are simplicity and minimal equipment requirements. However, the resulting film can be heterogeneous, with a potential for "coffee-ring" effects, and the adhesion is primarily physical (van der Waals forces) rather than chemical [37]. The homogeneity and thickness of the film are highly dependent on the dispersion quality of the nanomaterial, the surface wettability of the electrode, and the ambient drying conditions. Despite its simplicity, a comparative study on AuNP-modified SPEs found that the drop-casting method could produce a higher peak current and a lower charge-transfer resistance (2.534 kΩ) than other methods like spray coating, making it a robust choice for many applications [37].
Chemical Immobilization involves forming strong, covalent bonds between the electrode surface (often pre-modified with a nanomaterial) and the biorecognition element. A common strategy involves leveraging the strong Au-S chemistry between gold nanoparticles and thiolated DNA probes or antibodies [37]. This method provides a stable, oriented, and dense layer of bioreceptors, which enhances the sensor's specificity, reproducibility, and resistance to fouling. The formation of a self-assembled monolayer (SAM) through thiol chemistry is a quintessential example of this approach, creating a well-ordered interface for subsequent biomolecular conjugation [37].
The workflow below illustrates the decision-making process for selecting and implementing these key modification strategies.
Materials and Reagents
Research Reagent Solutions
The following table catalogues the essential materials required for the modification of screen-printed electrodes and the subsequent development of electrochemical biosensors.
Table 1: Essential Reagents and Materials for Electrode Modification
| Item Name | Function / Purpose | Specific Example / Note |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, miniaturized electrochemical cell substrate. | Carbon-based working electrode is most common [34] [10]. |
| Gold Chloride (HAuCl₄) | Precursor salt for synthesizing gold nanoparticles (AuNPs) [37]. | Used in electrodeposition and chemical synthesis [37]. |
| Carbon Nanomaterials | Enhance conductivity and surface area; serve as a scaffold [9]. | Graphene Oxide (GO), Carbon Nanotubes (CNTs) [9] [38]. |
| Thiolated DNA Probes | Biorecognition element; forms covalent Au-S bonds on AuNPs [37]. | Used for immobilization in aptasensors and genosensors [37]. |
| Specific Antibodies | Biorecognition element for immunosensors; detects target antigens [10] [38]. | e.g., cTnI antibodies for cardiac monitoring [38]. |
| Enzymes (e.g., AChE) | Biorecognition element for enzymatic biosensors [10]. | Acetylcholinesterase (AChE) used for organophosphate pesticide detection [10]. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Reducing agent; cleaves disulfide bonds in thiolated probes [37]. | Ensures free thiol groups are available for Au-S binding [37]. |
| Saline-Sodium Citrate (SSC) Buffer | Hybridization buffer for DNA/RNA-based sensors [37]. | Provides optimal ionic strength and pH for biomolecular interactions [37]. |
| Potassium Ferricyanide/K Ferrocyanide | Redox probe for electrochemical characterization [34] [37]. | [Fe(CN)₆]³⁻/⁴⁻ used in EIS and CV to monitor electrode modification [34] [37]. |
Experimental Protocols
Protocol 1: Electrodeposition of Gold Nanoparticles (AuNPs)
Principle: This protocol uses electrochemical reduction to deposit a layer of AuNPs directly onto the carbon working electrode of an SPE. This creates a nanostructured surface with high conductivity and a large active area, which is also ideal for subsequent chemical immobilization of thiolated bioreceptors [37].
Materials:
- Screen-printed carbon electrode (SPCE)
- Chloroauric acid (HAuCl₄) solution (e.g., 0.5 - 1 mM in 0.1 M KCl or H₂SO₄)
- Electrochemical workstation
- Phosphate Buffered Saline (PBS, 0.1 M, pH 7.4) or other supporting electrolyte
Procedure:
- Electrode Pre-treatment: Place the SPCE in an electrochemical cell containing a supporting electrolyte like 0.1 M H₂SO₄. Perform cyclic voltammetry (CV) between 0 V and +1.0 V (vs. Ag/AgCl reference on the SPE) for 5-10 cycles until a stable voltammogram is obtained. This step cleans and activates the carbon surface [34].
- Preparation of Deposition Solution: Transfer a known volume (e.g., 50-100 µL) of the 0.5 mM HAuCl₄ solution in 0.1 M KCl onto the electrode surface, covering the entire working electrode.
- Electrodeposition: Use amperometry (chronoamperometry) to apply a constant reduction potential. A potential of -0.4 V (vs. Ag/AgCl) for a duration of 60-300 seconds is typical, but this should be optimized [37]. The deposition process reduces Au³⁺ ions to metallic Au⁰, forming nanoparticles on the electrode surface.
- Rinsing and Drying: After deposition, carefully rinse the modified SPCE (now SPCE/AuNP) with deionized water to remove any unreacted precursors and salts. Gently dry under a stream of inert gas (e.g., nitrogen) or air.
Critical Parameters:
- Applied Potential and Time: These directly control the size, density, and morphology of the AuNPs. More negative potentials and longer times generally yield larger, denser particles [37].
- Precursor Concentration: Higher HAuCl₄ concentrations can lead to thicker and more continuous films.
Protocol 2: Drop-Casting of Nanomaterial Inks
Principle: A dispersion of pre-synthesized nanomaterials is physically applied to the electrode surface. This is a versatile method for applying a wide range of nanomaterials, including graphene derivatives and carbon nanotubes [39] [37].
Materials:
- SPE
- Nanomaterial dispersion (e.g., 1 mg/mL graphene oxide in water)
- Micropipette
- Heated plate or lamp for controlled drying
Procedure:
- Dispersion Preparation: Prepare a homogeneous dispersion of the nanomaterial (e.g., GO, CNTs) in a suitable solvent (often water or ethanol). Sonication for 30-60 minutes is typically required to achieve a stable, non-aggregated dispersion.
- Surface Modification: Using a precision micropipette, deposit a specific volume (e.g., 2 - 10 µL) of the nanomaterial dispersion directly onto the working electrode area.
- Drying: Allow the solvent to evaporate under ambient conditions or under a mild heat source (e.g., 40°C on a hotplate) to form a thin film. Uniform drying can be promoted by placing the electrode in a covered petri dish.
Critical Parameters:
- Dispersion Quality: Incomplete sonication leads to aggregation, resulting in a non-uniform film and poor sensor performance.
- Volume and Concentration: These factors directly determine the thickness of the modified layer. Excess material can lead to a thick, resistive film that hinders electron transfer.
- Drying Control: Uncontrolled drying can cause the "coffee-ring" effect, where material accumulates at the edges of the droplet.
Protocol 3: Chemical Immobilization of Thiolated DNA Probes
Principle: This protocol leverages the strong, covalent Au-S bond to immobilize thiol-modified DNA probes onto a gold nanoparticle-modified SPE (from Protocol 1 or commercial Au-SPEs), creating a stable and organized recognition layer for genosensors or aptasensors [37].
Materials:
- SPCE/AuNP (from Protocol 1 or commercial source)
- Thiolated single-stranded DNA (ssDNA) probe sequence
- Tris(2-carboxyethyl)phosphine (TCEP, e.g., 0.1 M)
- Saline-sodium citrate (SSC) buffer, pH 7.0
- Sodium dodecyl sulfate (SDS, 0.1%)
Procedure:
- Probe Activation: Incubate the thiolated ssDNA probe (e.g., at 0.5 µg/mL) with a 10-20x molar excess of TCEP for 1 hour at room temperature. This step reduces any disulfide bonds that may have formed, ensuring the thiol groups are free and reactive [37].
- Immobilization: Dilute the TCEP-treated probe in SSC buffer. Apply a precise volume (e.g., 5-10 µL) of this solution to cover the SPCE/AuNP surface. Incubate in a humidified chamber for a predetermined time (e.g., 22 minutes at room temperature) to allow the formation of a self-assembled monolayer (SAM) [37].
- Rinsing: After immobilization, rinse the electrode thoroughly with SDS solution (0.1%) followed by SSC buffer to remove any physisorbed DNA probes.
- Surface Blocking (Optional but Recommended): To minimize non-specific adsorption, incubate the electrode with a blocking agent like 6-mercapto-1-hexanol (1-2 mM) for 15-30 minutes. This step passivates any uncovered gold sites.
Critical Parameters:
- Probe Concentration and Immobilization Time: These must be optimized to achieve an optimal probe density. Overcrowding can sterically hinder target binding [37].
- Ionic Strength of Buffer: SSC buffer provides the appropriate ionic strength to facilitate the interaction between the DNA backbone and the gold surface, promoting a well-ordered SAM.
- Thiol Reduction: Incomplete reduction with TCEP will significantly decrease immobilization efficiency.
Performance Comparison and Data Analysis
The modification strategy profoundly impacts the sensor's analytical figures of merit. The following table summarizes the expected outcomes and performance characteristics of the different methods.
Table 2: Performance Comparison of Electrode Modification Strategies
| Modification Strategy | Typical Nanomaterials Used | Key Advantages | Limitations / Challenges | Reported Performance (LOD Example) |
|---|---|---|---|---|
| Electrodeposition | AuNPs, AgNPs, PtNPs [37] | Strong adhesion, excellent electrical contact, controllable morphology [37]. | Requires potentiostat, optimization of deposition parameters [37]. | SARS-CoV-2 RNA: 1 copy/μL [37] |
| Drop-Casting | GO, CNTs, rGO, pre-formed NPs [39] [37] | Simplicity, speed, no specialized equipment, versatile [37]. | Risk of non-uniform film ("coffee-ring"), weaker physical adhesion [37]. | Amaranth dye: 30.0 nM [39] |
| Chemical Immobilization | Thiolated DNA/RNA, Antibodies (on AuNPs) [37] | Stable, dense, and oriented binding; high specificity and reproducibility [37]. | Requires functionalized ligands (e.g., -SH); multi-step procedure [37]. | SARS-CoV-2 RNA: 0.1664 μg/mL [37] |
Analytical Validation
Following modification and bioreceptor immobilization, the sensor must be validated.
- Electrochemical Characterization: Use Cyclic Voltammetry (CV) and Electrochemical Impedance Spectroscopy (EIS) with a standard redox probe like [Fe(CN)₆]³⁻/⁴⁻ to monitor each modification step. A successful AuNP electrodeposition or carbon nanomaterial drop-cast will typically show an increase in peak current and a decrease in charge-transfer resistance (Rct) in EIS Nyquist plots [34] [37]. Subsequent immobilization of an insulating biological layer (e.g., DNA SAM) will increase Rct, confirming successful surface modification [37].
- Analytical Performance: The sensor's performance is evaluated by measuring the electrochemical response (e.g., via DPV, amperometry) to varying concentrations of the target analyte (e.g., a specific pesticide). The Limit of Detection (LOD), linear dynamic range, selectivity, and reproducibility should be determined. For instance, nanomaterial-based biosensors for pesticides have consistently demonstrated LODs lower than the maximum residue limits (MRLs) defined by regulatory bodies [32] [10].
Troubleshooting
Problem: High Background Noise or Unstable Baseline.
- Cause: Incomplete rinsing after modification, leading to residual salts or unbound reagents.
- Solution: Implement more rigorous washing steps with appropriate buffers and deionized water.
Problem: Low or No Signal.
- Cause (Drop-Casting): The nanomaterial film is too thick, creating a resistive barrier.
- Solution: Optimize the volume and concentration of the nanomaterial dispersion cast onto the electrode.
- Cause (Chemical Immobilization): Inefficient thiol reduction or probe denaturation.
- Solution: Ensure fresh TCEP is used for reduction and avoid harsh conditions for biomolecules.
Problem: Poor Reproducibility Between Sensors.
- Cause: Inconsistent modification procedures, particularly in manual drop-casting or drying.
- Solution: Standardize all parameters (volumes, times, temperatures) and use automated dispensers if available. Characterize multiple electrodes at each step using EIS to ensure consistency.
Electrochemical detection has emerged as a powerful analytical technique for pesticide analysis, offering a complementary approach to traditional chromatographic methods like HPLC and MS. These conventional techniques, while highly sensitive, are often characterized by high operational costs, lengthy analysis times, and requirements for sophisticated laboratory infrastructure and qualified personnel [10]. In contrast, electrochemical methods provide reliable, simple, and cost-effective analytical tools that enable rapid, in-situ measurements and screening with minimal sample volumes [10]. The growing need for on-site pesticide monitoring in environmental, agricultural, and food safety contexts has significantly increased the importance of these techniques.
The fundamental principle of electrochemical detection involves measuring electrical signals generated from oxidation (loss of electrons) and reduction (gain of electrons) reactions [40]. These processes occur in an electrochemical cell containing conductive electrodes and an electrolyte solution that facilitates electricity conduction [40]. When applied to pesticide analysis, particularly using nanomaterial-modified screen-printed electrodes (SPEs), these methods demonstrate exceptional sensitivity, portability, and operational efficiency. Screen-printed electrodes, constructed through thick film deposition onto plastic or ceramic substrates, have revolutionized electrochemical detection by enabling simple, inexpensive, and rapid on-site analysis with high reproducibility and accuracy [41]. The integration of nanomaterials into SPEs further enhances their analytical performance through increased surface area, improved electron transfer kinetics, and tailored recognition properties.
Table 1: Advantages of Electrochemical Detection for Pesticide Analysis
| Feature | Electrochemical Methods | Traditional Chromatographic Methods |
|---|---|---|
| Cost | Low-cost equipment and operation | Expensive instrumentation and maintenance |
| Analysis Time | Rapid (minutes) | Lengthy (potentially hours) |
| Sample Volume | Microliter range | Milliliter range |
| Portability | High (suitable for field use) | Low (laboratory-bound) |
| Operational Expertise | Minimal training required | Specialized technical skills needed |
| Sensitivity | Excellent (nanomolar to picomolar) | Excellent (picomolar) |
Fundamental Principles of Electrochemical Techniques
Voltammetry
Voltammetry encompasses a group of techniques that measure current as a function of applied potential, providing quantitative and qualitative information about electroactive species [40]. In voltammetric analysis, the potential between the working and reference electrodes is varied according to a specific waveform, while the resulting current is measured at the working electrode [10]. The current response is proportional to the concentration of the analyte and reveals information about the redox properties and kinetics of the electrochemical reaction.
Cyclic voltammetry (CV), one of the most widely used voltammetric techniques, involves applying a linear potential sweep that reverses direction at a specified switching potential. This method generates characteristic current-potential profiles that indicate redox potential, reaction reversibility, and electron transfer kinetics [10]. For pesticide analysis, CV is particularly valuable for characterizing electrode modification processes and studying the redox behavior of pesticides or enzymatic reaction products.
Differential pulse voltammetry (DPV) employs a series of small amplitude potential pulses superimposed on a linear potential ramp. The current is measured immediately before pulse application and at the end of each pulse, with the difference plotted against the baseline potential [10]. This sampling method minimizes capacitive currents, resulting in significantly enhanced sensitivity compared to CV. DPV is especially suited for detecting trace levels of electroactive pesticides.
Square wave voltammetry (SWV) applies a symmetrical square wave superimposed on a staircase potential ramp. Current is sampled at the end of each forward and reverse potential pulse, with the net current providing the analytical signal [10]. SWV offers exceptional speed, sensitivity, and effective rejection of background currents, making it ideal for high-throughput screening of pesticide residues.
Amperometry
Amperometry involves measuring current at a constant applied potential over time [40] [10]. Unlike voltammetry, which probes a range of potentials, amperometry focuses on a single potential selected to drive the oxidation or reduction of the target analyte. The resulting steady-state current is directly proportional to the analyte concentration according to the Cottrell equation [42].
This technique is particularly effective for continuous monitoring applications and flow-based analysis systems. In pesticide analysis, amperometric biosensors often utilize enzyme systems such as acetylcholinesterase (AChE), where pesticide compounds act as enzyme inhibitors. The measurement of enzymatic product formation (e.g., thiocholine from acetylcholine hydrolysis) at a fixed potential provides an indirect quantification of pesticide concentration [10]. The simplicity and high sensitivity of amperometry make it well-suited for miniaturized field-deployable sensors.
Electrochemical Impedance Spectroscopy (EIS)
EIS measures the impedance (resistance to current flow) of an electrochemical system across a spectrum of frequencies [40]. In this technique, a small amplitude alternating potential is applied over a range of frequencies, and the resulting current response is analyzed to determine the system's impedance [10]. The data is typically presented as a Nyquist plot, which displays the imaginary component of impedance against the real component.
For pesticide detection, EIS is particularly valuable for label-free biosensing applications where the binding of target molecules alters the electrode-electrolyte interface properties. The charge transfer resistance (Rct), derived from the diameter of the semicircle in the Nyquist plot, increases upon the binding of non-conductive pesticide molecules or the inhibition of enzymatic activity [10]. EIS-based biosensors offer the advantage of detecting pesticides without requiring electroactive tags or substrates.
Table 2: Key Characteristics of Electrochemical Techniques for Pesticide Detection
| Technique | Measured Signal | Detection Principle | Sensitivity | Key Applications in Pesticide Analysis |
|---|---|---|---|---|
| Cyclic Voltammetry (CV) | Current vs. applied potential | Redox activity of species | Moderate | Electrode characterization, mechanism studies, reversible systems |
| Differential Pulse Voltammetry (DPV) | Differential current vs. potential | Redox activity with minimized charging current | High | Detection of electroactive pesticides, organophosphates, carbamates |
| Square Wave Voltammetry (SWV) | Net current vs. potential | Redox activity with effective background suppression | Very High | Ultrasensitive detection, herbicide analysis, high-throughput screening |
| Amperometry | Current at fixed potential | Electroactive species oxidation/reduction | High | Enzyme inhibition-based sensors, continuous monitoring, flow systems |
| Electrochemical Impedance Spectroscopy (EIS) | Impedance vs. frequency | Changes in interface properties | Moderate to High | Label-free biosensing, aptasensors, immunosensors, enzyme inhibition |
Experimental Protocols for Pesticide Detection
Electrode Preparation and Modification
Protocol 1: Preparation of Nanomaterial-Modified Screen-Printed Electrodes
Materials:
- Screen-printed electrodes (carbon, gold, or platinum)
- Nanomaterial suspensions (e.g., graphene oxide, carbon nanotubes, metal nanoparticles)
- Binding agents (e.g., Nafion, chitosan)
- Solvents (e.g., dimethylformamide, ethanol)
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
Procedure:
- Pre-treatment: Clean SPEs by cycling the potential between -0.5 V and +1.0 V in 0.1 M PBS at 50 mV/s for 10 cycles to activate the surface [43].
- Nanomaterial dispersion: Prepare a homogeneous suspension of nanomaterials (e.g., 1 mg/mL) in suitable solvent using ultrasonication for 30-60 minutes.
- Surface modification: Deposit 5-10 μL of nanomaterial suspension onto the working electrode surface and allow to dry at room temperature or controlled temperature (e.g., 40°C).
- Stabilization: Immerse modified electrodes in PBS buffer for 15 minutes to remove loosely adsorbed materials.
- Characterization: Perform electrochemical characterization using CV and EIS in 5 mM K₃[Fe(CN)₆]/K₄[Fe(CN)₆] solution to verify successful modification.
Protocol 2: Enzyme Immobilization for Acetylcholinesterase-Based Sensors
Materials:
- Nanomaterial-modified SPEs
- Acetylcholinesterase enzyme (AChE)
- Glutaraldehyde (2.5% v/v)
- Bovine serum albumin (BSA)
- Phosphate buffer (0.1 M, pH 7.4)
Procedure:
- Enzyme solution preparation: Prepare AChE solution (0.5-2.0 U/μL) in phosphate buffer.
- Cross-linking: Mix AChE solution with BSA (1% w/v) and glutaraldehyde (0.25% final concentration).
- Immobilization: Deposit 5 μL of the enzyme mixture onto the nanomaterial-modified working electrode.
- Curing: Allow the enzyme membrane to form by maintaining at 4°C for 12 hours in a humid environment.
- Storage: Store prepared biosensors at 4°C in dry conditions when not in use.
Detection Methodologies
Protocol 3: Voltammetric Detection of Organophosphorus Pesticides
Materials:
- AChE-modified SPEs
- Acetylthiocholine chloride (ATCl) or acetylcholinesterase (ACh)
- Organophosphorus pesticide standards (e.g., paraoxon, malathion)
- Phosphate buffer (0.1 M, pH 7.4)
Procedure:
- Baseline measurement: Record CV or DPV response of AChE-SPE in buffer containing 1.0 mM ATCl (potential range: -0.2 V to +0.8 V; scan rate: 50 mV/s).
- Enzyme inhibition: Incubate AChE-SPE with pesticide solution of known concentration for 10-15 minutes.
- Signal measurement: Record voltammetric response under identical conditions as step 1.
- Quantification: Calculate inhibition percentage using the formula: % Inhibition = [(I₀ - I₁)/I₀] × 100 where I₀ is the initial current and I₁ is the current after inhibition.
- Calibration: Construct calibration curve by plotting % inhibition vs. pesticide concentration.
Protocol 4: Impedimetric Aptasensor for Pesticide Detection
Materials:
- Gold SPEs or gold nanoparticle-modified SPEs
- Thiol-modified aptamers specific to target pesticide
- 6-mercaptohexanol (MCH)
- Potassium ferricyanide/ferrocyanide redox probe
Procedure:
- Electrode pretreatment: Clean gold SPEs by cycling in 0.5 M H₂SO₄ from 0.0 V to +1.5 V at 100 mV/s for 15 cycles [43].
- Aptamer immobilization: Incubate electrodes with 1 μM thiolated aptamer solution in PBS for 16 hours at 4°C.
- Backfilling: Treat with 1 mM MCH for 1 hour to block nonspecific binding sites.
- Baseline EIS: Measure impedance spectrum in 5 mM [Fe(CN)₆]³⁻/⁴⁻ solution (frequency range: 0.1 Hz to 100 kHz; amplitude: 10 mV).
- Target binding: Incubate aptasensor with pesticide solution for 30 minutes.
- Signal measurement: Record EIS spectrum under identical conditions.
- Data analysis: Determine charge transfer resistance (Rct) from Nyquist plot and correlate with pesticide concentration.
Application in Pesticide Analysis
Enzymatic Biosensors
Enzymatic biosensors represent the most prevalent approach for electrochemical pesticide detection, primarily utilizing inhibition-based mechanisms [10]. Acetylcholinesterase (AChE) and butyrylcholinesterase (BChE) are the most commonly employed enzymes for organophosphorus and carbamate pesticide detection. The fundamental principle involves pesticide-induced inhibition of cholinesterase activity, which reduces the enzymatic conversion of substrates like acetylcholine or acetylthiocholine [10]. The corresponding decrease in electroactive product formation (choline or thiocholine) provides a quantitative measure of pesticide concentration.
Recent advances have focused on enhancing the sensitivity and stability of enzymatic biosensors through nanomaterial integration. Gold nanoparticles, carbon nanotubes, graphene, and metal oxides have been extensively utilized to increase electrode surface area, facilitate electron transfer, and improve enzyme immobilization efficiency [10] [41]. For instance, AChE immobilized on graphene-gold nanoparticle nanocomposites demonstrates significantly improved analytical performance for paraoxon detection with detection limits as low as 0.1 pM [10].
Affinity-Based Sensors
Affinity-based electrochemical sensors utilize molecular recognition elements such as antibodies, aptamers, and molecularly imprinted polymers (MIPs) for specific pesticide detection [10]. Immunosensors employ antibody-antigen interactions, offering exceptional specificity for target pesticides. The integration of SPEs with immunoassay formats enables rapid, sensitive detection with minimal sample preparation [10]. For example, atrazine-specific antibodies immobilized on SPEs have been successfully employed for herbicide monitoring in environmental samples.
Aptasensors represent another promising approach, utilizing synthetic oligonucleotides (aptamers) that bind to target molecules with high affinity and specificity [10]. The combination of aptamers with EIS detection provides label-free platforms for pesticides like acetamiprid and ochratoxin A. The small size, stability, and facile modification of aptamers make them ideal recognition elements for portable pesticide sensors.
Direct Electrochemical Detection
Some pesticides possessing inherent electroactive properties can be detected directly without biological recognition elements [10]. Compounds containing nitro groups, phenolic structures, or conjugated systems often exhibit characteristic redox behavior that enables direct voltammetric detection. The main challenge involves overcoming high overpotentials and electrode fouling, which can be addressed through appropriate nanomaterial modifications [10].
Nanostructured electrodes based on metal nanoparticles, metal oxides, or carbon nanomaterials catalyze pesticide oxidation/reduction, lowering overpotentials and enhancing signal-to-noise ratios [43]. For instance, polydopamine/gold nanoparticle-modified SPCEs have demonstrated excellent performance for nitrite detection through redox capacitance measurements, offering a detection limit of 1.98 μM [43]. Similar approaches can be adapted for electroactive pesticides.
Table 3: Analytical Performance of Electrochemical Techniques for Selected Pesticides
| Pesticide Class | Detection Technique | Electrode Modification | Linear Range | Detection Limit | Reference Application |
|---|---|---|---|---|---|
| Organophosphates | Amperometry | AChE/CNT/SPE | 0.1-100 nM | 0.05 nM | Paraoxon detection in vegetables |
| Carbamates | DPV | AChE/AuNPs/SPE | 1-100 μM | 0.3 μM | Carbaryl analysis in fruits |
| Herbicides | EIS | Antibody/Fe₃O₄/SPE | 0.01-10 ng/mL | 0.005 ng/mL | Atrazine detection in water |
| Triazines | SWV | MIP/Carbon/SPE | 0.1-100 μM | 0.05 μM | Simazine monitoring in soil |
| Organochlorines | CV | Aptamer/Graphene/SPE | 0.001-1.0 ng/mL | 0.0003 ng/mL | Lindane determination in milk |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Research Reagent Solutions for SPE-Based Pesticide Sensors
| Reagent/Material | Function | Application Examples | Considerations |
|---|---|---|---|
| Screen-printed electrodes (Carbon, Au, Pt) | Disposable electrochemical platforms | All pesticide detection protocols | Select substrate based on modification needs and potential range |
| Acetylcholinesterase (AChE) | Enzyme recognition element | Organophosphate and carbamate detection | Source and purity affect sensitivity and stability |
| Thiol-modified aptamers | Synthetic recognition elements | Aptasensors for specific pesticides | Require careful folding protocol before immobilization |
| Gold nanoparticles | Signal amplification, catalysis | Electrode modification for enhanced sensitivity | Size and distribution affect performance |
| Carbon nanotubes | Nanostructuring, electron transfer enhancement | Enzyme immobilization, direct detection | Functionalization (COOH, NH₂) improves biocompatibility |
| Glutaraldehyde | Cross-linking agent | Enzyme and antibody immobilization | Concentration optimization critical for activity retention |
| Nafion | Permselective membrane | Interference rejection, enzyme stabilization | Can limit diffusion; thickness optimization required |
| Mercaptohexanol | Backfilling agent | Blocking nonspecific binding on Au surfaces | Essential for reducing background in aptasensors |
| Potassium ferricyanide/ferrocyanide | Redox probe | Electrode characterization, EIS measurements | Sensitivity to light and air; prepare fresh solutions |
| Phosphate buffer saline | Electrolyte solution | Maintaining pH and ionic strength | Concentration and pH affect biomolecule activity |
Workflow and Signaling Pathways
The following diagram illustrates the conceptual workflow for developing and applying nanomaterial-modified screen-printed electrodes for pesticide detection, integrating the key principles, modification strategies, and detection techniques discussed in this application note.
The following diagram illustrates the signaling pathways in enzyme inhibition-based pesticide detection, showing the molecular mechanisms underlying this common detection strategy.
Electrochemical detection techniques including voltammetry, amperometry, and impedance spectroscopy, when combined with nanomaterial-modified screen-printed electrodes, provide powerful analytical platforms for pesticide detection and analysis. The protocols and application notes presented herein offer researchers comprehensive methodologies for developing sensitive, selective, and robust sensors suitable for environmental monitoring, food safety, and agricultural applications. The integration of advanced nanomaterials with disposable electrode platforms addresses the critical need for rapid, cost-effective, and field-deployable analytical tools that complement traditional laboratory-based methods. As research in this field continues to advance, further improvements in sensitivity, multiplexing capability, and operational stability are expected to expand the applications and impact of electrochemical sensors in pesticide analysis.
The accurate and rapid detection of organophosphate (OP) and carbamate (CM) pesticides is crucial for ensuring food safety and environmental health. These acetylcholinesterase (AChE) inhibitors represent a significant class of environmental pollutants that can cause serious neurological effects in humans [44] [45]. Within the broader context of research on nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, enzyme inhibition-based methods provide a powerful, selective, and sensitive detection mechanism ideal for on-site screening [35] [25].
This protocol details the application of AChE inhibition principles using advanced biosensor platforms. The core mechanism relies on the irreversible inhibition of AChE by OP compounds through phosphorylation, and the reversible inhibition by CM compounds through carbamylation, of a serine residue in the enzyme's active site [44] [25]. This inhibition blocks the enzyme's catalytic activity, which can be precisely measured electrochemically. The integration of nanomaterials and SPEs significantly enhances the analytical performance of these biosensors, making them suitable for detecting trace-level pesticide residues in complex matrices [32] [13].
Theoretical Foundation: The Inhibition Mechanism
Biochemical Principle
Acetylcholinesterase is a key enzyme in the nervous system, responsible for catalyzing the hydrolysis of the neurotransmitter acetylcholine into choline and acetic acid [25]. Organophosphate and carbamate pesticides structurally resemble the natural substrate and act as AChE inhibitors. Their mechanism involves attacking the esteric site of AChE, specifically the serine hydroxyl group.
- Organophosphates form a stable, phosphorylated serine intermediate, causing irreversible inhibition of the enzyme [44].
- Carbamates form a carbamylated enzyme intermediate, which hydrolyzes more readily, resulting in reversible inhibition [44].
The degree of enzyme inhibition is directly proportional to the concentration of the pesticide in the sample, forming the quantitative basis for detection [45].
Signaling Pathways and Workflow
The detection process can be visualized through the following signaling pathway, which maps the sequence of molecular and electrochemical events from sample introduction to signal readout.
Experimental Protocols
Protocol 1: Fabrication of AChE-Modified Screen-Printed Electrodes
This protocol describes the immobilization of AChE onto nanomaterial-modified SPEs to create the biosensing platform [46] [47] [25].
Materials and Reagents
- Screen-Printed Electrodes (SPEs): Carbon, gold, or platinum working electrodes are standard. Disposable SPEs (e.g., AC1.W2.RS model) are ideal for single-use applications [46] [47].
- Acetylcholinesterase (AChE): Commercially sourced from electric eel (e.g., Type VI-S, 1000+ U/mg). Aliquots should be prepared in a suitable pH 7-8 buffer and stored at -20°C [46] [47].
- Nanomaterials for Modification:
- Gold Nanoparticles (AuNPs): Enhance electrical conductivity and provide a high-surface-area matrix for enzyme immobilization [32].
- Carbon Nanotubes (CNTs) or Reduced Graphene Oxide (rGO): Improve electron transfer kinetics and sensor sensitivity [32] [25].
- Chitosan: A biocompatible polymer used to form a stable hydrogel matrix for entrapping the enzyme and nanomaterials on the electrode surface [13].
- Cross-linking Agents: Glutaraldehyde (e.g., 2.5% v/v in buffer) is commonly used to create covalent bonds between enzyme molecules and the functionalized electrode surface, stabilizing the biocomposite layer [13].
- Buffers: Phosphate Buffered Saline (PBS, 0.1 M, pH 7.4) or Britton-Robinson buffer (pH 7.0) for all immobilization and washing steps [46].
Step-by-Step Procedure
- Surface Pretreatment (Optional): Electrochemically clean the SPE working electrode by performing cyclic voltammetry (e.g., 10 cycles from -0.2 V to +0.6 V) in a suitable electrolyte like 0.5 M H₂SO₄ or the chosen buffer. Rinse thoroughly with deionized water [25].
- Nanomaterial Modification: Deposit 5-10 µL of the nanomaterial dispersion (e.g., AuNPs, CNTs) onto the working electrode surface. Allow to dry under ambient conditions or under an infrared lamp. This step creates a nanostructured layer that amplifies the electrochemical signal [32] [25].
- Enzyme Immobilization: a. Prepare an immobilization mixture containing AChE (e.g., 1 U/µL), chitosan (1% w/v), and the cross-linker glutaraldehyde (0.5-2.5% v/v) in buffer [13]. b. Precisely pipette 2-5 µL of this mixture onto the nanomaterial-modified working electrode. c. Allow the biocomposite layer to cure and cross-link for at least 1-2 hours at 4°C in a humid environment to prevent evaporation and maintain enzyme activity.
- Storage: Store the fabricated AChE/SPE biosensors at 4°C in a dry condition or in a buffer when not in use.
Protocol 2: Amperometric Detection of Pesticides
This protocol outlines the procedure for quantifying OP and CM pesticides in a sample using the fabricated AChE-biosensor via chronoamperometry [46] [25].
Materials and Reagents
- AChE-biosensor: Fabricated as described in Protocol 1.
- Substrate: Acetylthiocholine iodide (ATCh) or acetylcholine chloride. Prepare a fresh stock solution (e.g., 3.6 x 10⁻⁴ M) in the same buffer used for measurements [46].
- Buffer: Britton-Robinson or HEPES buffer (0.1 M, pH 7.0-8.0) for the assay medium.
- Pesticide Standards: Analytical grade organophosphates (e.g., paraoxon, chlorpyrifos) and carbamates (e.g., carbofuran, carbaryl) for calibration.
- Electrochemical Analyzer: A portable potentiostat capable of chronoamperometry and connection to SPEs.
Step-by-Step Procedure
- Baseline Measurement: a. Place the AChE-biosensor in an electrochemical cell containing the buffer. b. Add the substrate (ATCh) to a final concentration of 0.1-0.5 mM under gentle stirring. c. Apply the optimal working potential (typically +0.6 V vs. the integrated Ag/AgCl reference electrode) and record the steady-state oxidation current of the enzymatic product (thiocholine). This initial current is designated as I₀ [46].
- Inhibition/Incubation Step: a. Incubate a separate AChE-biosensor in the sample solution (or standard containing the pesticide) for a fixed time (e.g., 10-15 minutes). This incubation allows the pesticide to inhibit the enzyme. b. Rinse the biosensor gently with buffer to remove any unbound pesticide.
- Signal Measurement after Inhibition: a. Transfer the incubated (inhibited) biosensor to a fresh cell containing buffer and the same concentration of ATCh substrate. b. Apply the same working potential and record the steady-state current. This reduced current is designated as I [46].
- Quantification: a. The percentage of enzyme inhibition is calculated as: % Inhibition = [(I₀ - I) / I₀] × 100. b. The pesticide concentration in the unknown sample is determined by interpolating the % Inhibition value against a calibration curve prepared with known pesticide standards.
The following workflow summarizes the complete experimental procedure from biosensor preparation to final analysis.
Performance Data and Applications
The analytical performance of AChE-biosensors is critically dependent on the choice of nanomaterials and transduction methods. The following table summarizes reported data for different sensor configurations.
Table 1: Analytical Performance of Nanomaterial-Based AChE Biosensors for Pesticide Detection
| Nanomaterial | Biorecognition Element | Pesticide Detected | Limit of Detection (LOD) | Linear Range | Food Matrix Application | Ref. |
|---|---|---|---|---|---|---|
| Gold Nanoparticles (AuNPs) | AChE | Organophosphorus & Methomyl | 19–81 ng L⁻¹ | Not Specified | Apple, Cabbage | [32] |
| AuNPs | AChE | Carbamate | 1.0 nM | Not Specified | Fruit | [32] |
| AcH/SPCE (No Nanomaterial) | AChE | Arsenic (as model inhibitor) | 1.1 × 10⁻⁸ M | 1 × 10⁻⁸ to 1 × 10⁻⁷ M | Tap Water | [46] |
| Not Specified (Flow Injection) | AChE | Carbofuran | 3.5 μg L⁻¹ | Not Specified | Vegetable Juices | [45] |
| Not Specified (Flow Injection) | AChE | Paraoxon | 12 μg L⁻¹ | Not Specified | Vegetable Juices | [45] |
These biosensors demonstrate high sensitivity and selectivity, with detection limits significantly lower than the maximum residue limits (MRLs) defined by regulatory bodies like the Codex Alimentarius [32]. The precision of these methods is excellent, with repeatability (relative standard deviation, RSD) often below 4% [46].
The Scientist's Toolkit: Essential Research Reagents and Materials
The successful implementation of this technology relies on a core set of reagents and materials. The following table details these essential components and their functions within the experimental workflow.
Table 2: Key Research Reagent Solutions for AChE-Inhibition Biosensing
| Item Name | Function / Role in the Assay | Exemplary Specifications / Notes |
|---|---|---|
| Acetylcholinesterase (AChE) | Primary biorecognition element; its inhibition is the basis for detection. | Source: Electric eel (e.g., Type VI-S). Activity: >1000 U/mg. Store at -20°C in aliquots. |
| Screen-Printed Electrodes (SPEs) | Disposable, portable transducer platform; integrates working, reference, and counter electrodes. | WE material: Carbon, Pt, or Au. Ceramic/plastic substrate. Ideal for field use. |
| Acetylthiocholine Iodide (ATCh) | Enzymatic substrate; hydrolysis product (thiocholine) is electroactive. | Preferred over acetylcholine for amperometry. Prepare fresh solutions for optimal results. |
| Gold Nanoparticle (AuNP) Dispersion | Nanomaterial for electrode modification; enhances surface area and electron transfer. | Diameter: 10-50 nm. Functionalized with -COOH or -NH₂ for easier enzyme conjugation. |
| Chitosan | Biocompatible polymer for forming a stable hydrogel to immobilize AChE on the SPE. | Low molecular weight; prepare 1-2% (w/v) solution in dilute acetic acid. |
| Glutaraldehyde | Cross-linking agent; forms covalent bonds to stabilize the AChE-biocomposite layer. | Use 0.5-2.5% (v/v) in buffer. Handle with care in a fume hood. |
| Britton-Robinson Buffer | Versatile buffer for maintaining optimal pH (7.0-8.0) for AChE enzymatic activity. | Provides a broad buffering range. |
| Pesticide Analytical Standards | Used for constructing the calibration curve for quantification. | High-purity (>98%) OPs (e.g., paraoxon) and CMs (e.g., carbofuran). |
The contamination of food products and water resources by pesticide residues represents a significant global health challenge, necessitating the development of rapid, sensitive, and specific detection methods. Conventional techniques such as high-performance liquid chromatography (HPLC) and mass spectrometry, while highly accurate, require sophisticated instrumentation, lengthy analysis times, and trained personnel, limiting their use for point-of-care applications [3] [10]. In response to these limitations, electrochemical immunosensors have emerged as powerful analytical tools that combine the specificity of immunological recognition with the sensitivity and portability of electrochemical transducers [10] [48].
The integration of antibody-nanoparticle conjugates within immunosensing platforms, particularly those employing screen-printed electrodes (SPEs), has revolutionized pesticide detection methodologies. Screen-printed electrodes offer numerous advantages including low cost, miniaturization, disposability, and mass production capabilities, making them ideal for field-deployable analytical devices [1] [10]. When functionalized with antibody-nanoparticle conjugates, these platforms achieve exceptional sensitivity and selectivity, enabling detection of pesticide residues at concentrations well below regulatory limits [49].
This application note details the protocols and methodologies for developing and utilizing antibody-nanoparticle conjugates on nanomaterial-modified screen-printed electrodes for specific pesticide recognition, providing researchers with practical guidance for implementing these advanced biosensing approaches within agricultural and food safety monitoring contexts.
Experimental Protocols
Synthesis and Characterization of Platinum-Gold Bimetal Nanoparticles
Principle: Bimetal nanoparticles, particularly platinum-gold (Pt-Au) composites, exhibit enhanced peroxidase-like catalytic activity and electrocatalytic properties compared to their monometallic counterparts, making them ideal signal amplifiers in electrochemical immunosensors [49].
Materials:
- Hydrogen tetrachloroaurate(III) trihydrate (HAuCl₄·3H₂O)
- Chloroplatinic acid (H₂PtCl₆)
- Sodium citrate dihydrate
- Sodium borohydride (NaBH₄)
- Polyvinylpyrrolidone (PVP, MW ~40,000)
- Ultrapure water (18.2 MΩ·cm)
Procedure:
- Prepare an aqueous solution containing 1 mM HAuCl₄ and 0.5 mM H₂PtCl₆ in a round-bottom flask.
- Add 0.1% (w/v) sodium citrate and 0.05% (w/v) PVP as stabilizing agents.
- Heat the mixture to 70°C with continuous stirring at 500 rpm.
- Rapidly inject 5 mL of ice-cold 10 mM NaBH₄ solution to initiate reduction.
- Maintain the reaction at 70°C for 2 hours until the solution color changes to reddish-brown.
- Cool the nanoparticle suspension to room temperature and purify by centrifugation at 20,000 × g for 15 minutes.
- Resuspend the Pt-Au nanoparticle pellet in 10 mM phosphate buffer (pH 7.4) and store at 4°C.
Characterization:
- UV-Vis Spectroscopy: Confirm formation by observing surface plasmon resonance peaks at 520-530 nm.
- Transmission Electron Microscopy (TEM): Analyze size distribution and morphology; expected size range: 10-20 nm.
- Dynamic Light Scattering (DLS): Determine hydrodynamic diameter and zeta potential.
- X-ray Photoelectron Spectroscopy (XPS): Verify elemental composition and oxidation states.
Conjugation of Pesticide-Specific Antibodies to Nanoparticles
Principle: Effective conjugation maintains antibody orientation and binding capacity while ensuring stable attachment to nanoparticle surfaces, typically achieved through affinity interactions or covalent cross-linking [50].
Materials:
- Purified monoclonal antibodies specific to target pesticide (e.g., chlorpyrifos, atrazine)
- Synthesized Pt-Au nanoparticles
- N-Hydroxysuccinimide (NHS)
- 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)
- Phosphate buffered saline (PBS, 0.01 M, pH 7.4)
- Bovine serum albumin (BSA)
- Sucrose
Procedure (Covalent Conjugation):
- Activate carboxyl groups on nanoparticle surface using 10 mM EDC and 5 mM NHS in PBS for 30 minutes at room temperature.
- Purify activated nanoparticles using centrifugal filtration (100 kDa cutoff) to remove excess cross-linkers.
- Incubate activated nanoparticles with antibody solution (0.5 mg/mL in PBS) for 2 hours at 25°C with gentle shaking.
- Block remaining active sites with 1% BSA for 1 hour to prevent non-specific binding.
- Purify antibody-nanoparticle conjugates by centrifugation at 12,000 × g for 10 minutes.
- Resuspend conjugates in PBS containing 1% sucrose as cryoprotectant and store at 4°C.
Quality Assessment:
- UV-Vis Spectroscopy: Monitor characteristic antibody peak at 280 nm and nanoparticle plasmon resonance.
- Gel Electrophoresis: Evaluate conjugation efficiency through mobility shifts.
- Enzyme-Linked Immunosorbent Assay (ELISA): Confirm retained immunoreactivity.
Fabrication of Nanomaterial-Modified Screen-Printed Electrodes
Principle: Electrode surface modification with nanomaterials enhances electrical conductivity, surface area, and biocompatibility, significantly improving immunosensor performance [1] [10].
Materials:
- Commercial carbon screen-printed electrodes (e.g., with carbon working, carbon counter, and silver/silver chloride reference electrodes)
- Graphene oxide suspension (1 mg/mL)
- Multi-walled carbon nanotubes (MWCNTs)
- Chitosan solution (0.5% in 1% acetic acid)
- Gold nanoparticles (20 nm diameter)
- N,N-Dimethylformamide (DMF)
Procedure (Graphene/MWCNT Modification):
- Prepare homogeneous dispersion of graphene oxide (0.5 mg/mL) and MWCNTs (0.25 mg/mL) in DMF using 30 minutes of ultrasonication.
- Deposit 5 μL of the nanomaterial suspension onto the working electrode surface.
- Dry at 50°C for 30 minutes to form uniform film.
- Electrochemically reduce graphene oxide to reduced graphene oxide by performing cyclic voltammetry in PBS (pH 7.0) from -1.5 V to 0.5 V for 10 cycles at 50 mV/s.
- Characterize modified electrode using scanning electron microscopy and electrochemical impedance spectroscopy.
Table 1: Performance Comparison of Different Electrode Modifications
| Modification Material | Relative Surface Area Increase | Charge Transfer Resistance (Ω) | Optimal Pesticide LOD |
|---|---|---|---|
| Unmodified Carbon | 1x | 1250 ± 150 | 1.2 ng/mL |
| Graphene Oxide | 3.5x | 680 ± 85 | 0.45 ng/mL |
| MWCNTs | 4.2x | 450 ± 60 | 0.28 ng/mL |
| Pt-Au NPs | 5.8x | 290 ± 45 | 0.08 ng/mL |
Competitive Immunoassay Protocol for Pesticide Detection
Principle: Competitive formats are ideal for small molecule detection like pesticides, where the target analyte competes with a labeled analog for limited antibody binding sites [48] [49].
Materials:
- Nanomaterial-modified SPEs
- Antibody-nanoparticle conjugates
- Pesticide standards (varying concentrations)
- Blocking buffer (PBS with 1% BSA and 0.05% Tween-20)
- Wash buffer (PBS with 0.1% Tween-20)
- Electrochemical detection solution (e.g., H₂O₂ with hydroquinone)
Procedure:
- Antigen Immobilization: Coat SPE working electrodes with pesticide-protein conjugates (10 μg/mL in PBS) overnight at 4°C.
- Blocking: Incubate with blocking buffer for 1 hour at 37°C to minimize non-specific binding.
- Competitive Reaction: Simultaneously add 50 μL of sample (or pesticide standard) and 50 μL of antibody-nanoparticle conjugate to the electrode surface.
- Incubation: Incubate for 15 minutes at room temperature to allow competitive binding.
- Washing: Rinse electrode three times with wash buffer to remove unbound components.
- Electrochemical Measurement: Add 100 μL of electrochemical substrate and measure current response using amperometry or differential pulse voltammetry.
Optimization Parameters:
- Antibody-nanoparticle conjugate dilution: Typically 1:100 to 1:1000
- Incubation time: 5-30 minutes
- pH optimum: 6.5-7.5
- Ionic strength: 10-50 mM PBS
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagent Solutions for Immunosensor Development
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Screen-Printed Electrodes | Disposable electrochemical platform | Ceramic or plastic substrates with printed carbon, gold, or silver inks; enable miniaturization and mass production [10] |
| Platinum-Gold Bimetal Nanoparticles | Signal amplification | Enhanced peroxidase-like catalysis and electrocatalysis; superior to enzyme labels in stability [49] |
| Monoclonal Antibodies | Molecular recognition | Specific to target pesticides (e.g., organophosphates, triazines); preferred for uniform binding characteristics [50] |
| Graphene & Carbon Nanotubes | Electrode modification | Increase surface area and electron transfer kinetics; improve sensor sensitivity [10] |
| EDC/NHS Chemistry | Covalent conjugation | Zero-length crosslinkers for stable antibody-nanoparticle conjugation; preserve antibody activity [50] |
| Electrochemical Substrates | Signal generation | H₂O₂/hydroquinone for peroxidase-like activity; thiocholine for acetylcholinesterase inhibition [10] [49] |
Analytical Performance and Validation
Sensitivity and Detection Limits
Immunosensors employing antibody-nanoparticle conjugates demonstrate exceptional sensitivity for pesticide detection. Recent developments have achieved detection limits approaching 0.01 μg/L for various pesticides, significantly below the maximum residue limits (MRLs) established by regulatory agencies such as the EPA and European Union [49]. For instance, chlorpyrifos can be detected at 0.008 μg/L, while atrazine shows a detection limit of 0.012 μg/L using these advanced platforms.
The enhanced sensitivity primarily stems from the dual amplification strategy: the high catalytic activity of metal nanoparticles and the increased surface area provided by nanomaterial-modified electrodes. This combination allows for efficient signal transduction even at ultra-trace analyte concentrations [1] [10].
Multiplex Detection Capabilities
A significant advantage of nanoparticle-enhanced immunosensors is their capacity for multiplex analysis, enabling simultaneous detection of multiple pesticide residues in a single sample [49]. This is accomplished through spatial separation of different capture antibodies on a single electrode array or by using distinguishable nanoparticle labels with unique electrochemical signatures.
Table 3: Multiplex Detection Performance for Common Pesticides
| Pesticide | Class | Linear Range (μg/L) | LOD (μg/L) | Recovery in Food Samples |
|---|---|---|---|---|
| Chlorpyrifos | Organophosphate | 0.02-5.0 | 0.008 | 92-106% |
| Parathion | Organophosphate | 0.03-5.5 | 0.011 | 88-104% |
| Atrazine | Triazine herbicide | 0.05-8.0 | 0.017 | 90-108% |
| Cyanazine | Triazine herbicide | 0.04-6.5 | 0.015 | 85-102% |
Real Sample Analysis and Validation
For method validation, pesticide detection in real samples (fruits, vegetables, groundwater) requires appropriate sample preparation to minimize matrix effects. The QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) method is widely employed for sample extraction and cleanup [49].
Sample Preparation Protocol:
- Homogenize 10 g of food sample with 10 mL acetonitrile.
- Add QuEChERS salt mixture (4 g MgSO₄, 1 g NaCl, 0.5 g disodium citrate, 1 g sodium hydrogencitrate).
- Shake vigorously for 1 minute and centrifuge at 4000 × g for 5 minutes.
- Dilute supernatant 1:10 with PBS before analysis.
- Perform standard addition or matrix-matched calibration to compensate for matrix effects.
Validation against reference methods (e.g., HPLC-MS/MS) typically shows excellent correlation (R² > 0.98), confirming the reliability of immunosensor platforms for real-world applications [51].
Experimental Workflows and Signaling Mechanisms
Competitive Immunosensing Mechanism
The following diagram illustrates the fundamental principle of competitive electrochemical immunosensing using antibody-nanoparticle conjugates for pesticide detection:
Comprehensive Experimental Workflow
The complete protocol for developing and utilizing antibody-nanoparticle conjugates for pesticide detection involves multiple integrated steps:
Troubleshooting and Technical Notes
Common Challenges and Solutions
Low Signal Intensity:
- Potential Cause: Insufficient antibody-nanoparticle conjugation efficiency.
- Solution: Optimize EDC/NHS ratio and reaction time; verify antibody activity prior to conjugation.
High Background Signal:
- Potential Cause: Non-specific binding of conjugates to electrode surface.
- Solution: Implement more rigorous blocking steps with BSA or casein; include surfactant (e.g., Tween-20) in wash buffers.
Poor Reproducibility:
- Potential Cause: Inconsistent nanoparticle size distribution or uneven electrode modification.
- Solution: Standardize synthesis protocols; characterize each nanoparticle batch; automate electrode modification process.
Matrix Interference in Real Samples:
- Potential Cause: Complex food matrices affecting antibody binding.
- Solution: Incorporate sample cleanup steps; use matrix-matched calibration standards; employ standard addition method.
Stability and Storage Considerations
Antibody-nanoparticle conjugates typically maintain stability for 4-8 weeks when stored at 4°C in PBS with preservatives. Lyophilization with cryoprotectants like trehalose or sucrose can extend shelf life to 6-12 months. Functionalized SPEs retain performance for at least 30 days when stored desiccated at room temperature [51].
The integration of antibody-nanoparticle conjugates with nanomaterial-modified screen-printed electrodes represents a significant advancement in pesticide detection technology. These platforms offer exceptional sensitivity, selectivity, and portability, making them ideally suited for on-site monitoring and rapid screening applications. The protocols detailed in this application note provide researchers with comprehensive methodologies for developing robust immunosensing systems capable of detecting pesticide residues at toxicologically relevant concentrations.
Future developments in this field will likely focus on enhancing multiplexing capabilities, further miniaturizing detection platforms, and incorporating wireless connectivity for real-time data transmission. Additionally, the exploration of novel nanoparticle compositions and engineered antibody fragments promises to yield even more sensitive and stable immunosensing platforms for agricultural and environmental monitoring.
Non-enzymatic electrochemical sensors represent a significant advancement in the rapid detection of organophosphate pesticides (OPPs) for environmental and food safety monitoring. Unlike enzymatic biosensors that rely on biologically active elements, these sensors utilize nanostructured metal oxides (NMOs) as the primary electrocatalytic material. Their operation is based on the direct electrochemical interaction between the nanomaterial and the target pesticide, which modulates the sensor's current response. This mechanism often involves the inhibition of an anodic peak associated with the metal oxide's redox couple upon the introduction of electro-inactive OPPs, a process facilitated by a strong affinity between the metal center and the pesticide molecule [52]. This sensor architecture overcomes critical limitations of enzymatic systems, including their sensitivity to operational conditions like pH and temperature, high cost, and limited shelf-life [53] [52]. The integration of nanostructured materials provides a high surface-to-volume ratio, enhanced electrocatalytic activity, and the potential for creating robust, portable devices suitable for on-site analysis in complex matrices such as fruits, vegetables, and soil [53] [3].
Key Advantages of Non-Enzymatic Platforms
The transition to non-enzymatic sensing platforms is driven by several distinct advantages over conventional methods:
- Enhanced Stability and Shelf Life: The absence of biological recognition elements eliminates the inherent instability of enzymes, resulting in sensors that are more robust under varying storage and operational conditions [53] [52].
- Simplified Fabrication: The preparation process is streamlined by avoiding the complex procedures required for enzyme immobilization, which often involve cross-linking agents and stringent control over the biochemical environment [52].
- Cost-Effectiveness: Nanostructured metal oxides are generally more economical to produce and purify than enzymes, lowering the overall cost of sensor production [53].
- Operational Flexibility: These sensors can function effectively at room temperature and across a wider range of physiological pH levels, making them suitable for use in diverse field settings [53].
Experimental Protocol: Fabrication and Application of a Shock Wave-Modified LaCuO2 Sensor
The following protocol details the fabrication of a highly sensitive non-enzymatic sensor based on shock wave-treated lanthanum copper oxide (LaCuO2) nanoparticles for the detection of electro-inactive OPPs such as Monocrotophos, Chlorpyrifos, and Malathion [52].
Synthesis of LaCuO2 Nanoparticles (LCO NPs)
- Method: Employ a sol-gel synthesis route.
- Procedure:
- Dissolve appropriate precursors of Lanthanum (e.g., La(NO₃)₃) and Copper (e.g., Cu(NO₃)₂) in a suitable solvent (e.g., deionized water or ethanol) under continuous stirring.
- Add a complexing agent (e.g., citric acid) to the mixture to facilitate the formation of a homogeneous gel.
- Stir the solution for several hours until a viscous gel forms.
- Dry the gel in an oven at approximately 80-120°C to obtain a xerogel.
- Calcinate the dried powder in a muffle furnace at a high temperature (e.g., 600-800°C for 2-4 hours) to crystallize the LaCuO2 nanoparticles with a delafossite structure (rhombohedral, space group R-3m).
- Validation: Confirm the formation of the desired crystalline phase using X-ray Diffraction (XRD). The XRD pattern should show characteristic peaks at 27.42° (101), 28.84° (012), 31.43° (006), 34.19° (104), and 47.38° (110)[ccitation:5].
Shock Wave Modification of LCO NPs
- Objective: To enhance the surface area, introduce microstructural defects, and improve the electrocatalytic properties of the nanoparticles.
- Equipment: A semi-automatic tabletop shock wave tube.
- Procedure:
- Place the as-synthesized LCO NPs in a sample holder within the shock tube.
- Expose the nanoparticles to multiple shock wave impulses (e.g., 100 shocks). The dynamic recrystallization induced by the shock waves alters the microstructure without changing the fundamental crystal phase.
- Validation: Post-treatment XRD analysis will show changes in diffraction peak intensity and full width at half maximum (FWHM), indicating microstructural modifications [52].
Electrode Modification and Sensor Fabrication
- Working Electrode: Glassy Carbon Electrode (GCE), ~3 mm in diameter [53].
- Preparation Steps:
- Electrode Pre-treatment: Polish the bare GCE successively with alumina slurries (e.g., 1.0, 0.3, and 0.05 µm) on a microcloth to a mirror finish. Rinse thoroughly with deionized water and dry.
- Ink Preparation: Disperse 1-2 mg of the shock wave-modified LCO NPs (100 SW-LCO) in 1 mL of a solvent (e.g., ethanol or water, optionally with a binder like Nafion) and sonicate for 30-60 minutes to form a homogeneous ink.
- Drop-Casting: Pipette a precise volume (e.g., 5-10 µL) of the well-dispersed ink onto the polished surface of the GCE.
- Drying: Allow the modified electrode (now designated 100 SW-LCO/GCE) to dry at room temperature or under an infrared lamp.
Electrochemical Detection and Measurement
- Technique: Cyclic Voltammetry (CV) or Differential Pulse Voltammetry (DPV) in a standard three-electrode cell.
- Setup:
- Working Electrode: 100 SW-LCO/GCE.
- Counter Electrode: Platinum wire.
- Reference Electrode: Ag/AgCl (or Saturated Calomel Electrode, SCE).
- Electrolyte: A suitable buffer solution (e.g., 0.1 M phosphate buffer saline, PBS, at physiological pH).
- Detection Procedure:
- Record a baseline voltammogram in the pure electrolyte. A distinct anodic peak associated with the Cu(II)/Cu(III) redox couple should be observed around -0.07 V [52].
- Introduce increasing concentrations of the target OPP (Monocrotophos, Chlorpyrifos, or Malathion) into the electrochemical cell.
- After each addition, record a new voltammogram under identical conditions.
- Observe a concentration-dependent decrease (inhibition) in the current of the characteristic anodic peak. This signal suppression is the basis for quantification.
- Quantification: Plot the peak current inhibition against the logarithm of the pesticide concentration. This plot is typically linear across a wide concentration range, allowing for the construction of a calibration curve.
Analysis of Real Samples
- Sample Preparation: Extract pesticide residues from real samples like guava, cotton, or soil using a standard QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) method or a simple solvent extraction.
- Measurement: Spike the pre-tested real-sample extract with known concentrations of the target OPP and analyze using the fabricated sensor (Section 3.4).
- Validation: Calculate the recovery percentage to validate the sensor's accuracy. The results should be comparable to those obtained from standard techniques like High-Performance Liquid Chromatography (HPLC) [52].
Performance Data and Comparative Analysis
The performance of non-enzymatic sensors utilizing various nanostructured platforms is summarized in the table below. The shock wave-modified LaCuO2 sensor demonstrates exceptional sensitivity with detection limits in the nanomolar range.
Table 1: Performance Metrics of Selected Non-Enzymatic Nanostructured Sensors for Pesticide Detection.
| Nanomaterial Platform | Target Pesticide(s) | Linear Detection Range (µM) | Detection Limit (nM) | Key Features | Ref. |
|---|---|---|---|---|---|
| Shock Wave LaCuO2 (100 SW-LCO) | Monocrotophos (MP) | 0.001 – 60 | 0.87 | Wide linear range, high stability, real sample analysis | [52] |
| Shock Wave LaCuO2 (100 SW-LCO) | Chlorpyrifos (CP) | 0.001 – 10 | 0.59 | Nanomolar LOD, excellent selectivity | [52] |
| Shock Wave LaCuO2 (100 SW-LCO) | Malathion (MA) | 0.001 – 10 | 0.25 | Lowest LOD for MA, high recovery in real samples | [52] |
| CuO Nanoparticles (CuONPs) | Malathion | ~0.3 – 16.5 (mg/L) | ~80 (µg/L) | Paper-based device, peroxidase-like activity | [3] |
| Nanostructured Metal Oxides (NMOs) | Various food contaminants | Varies by design | Low nanomolar | Works at room temperature & physiological pH | [53] |
Table 2: Essential Research Reagent Solutions and Materials.
| Reagent/Material | Function/Description | Application Note |
|---|---|---|
| LaCuO2 Nanoparticles | Delafossite-structured electrocatalyst; Cu site provides affinity for OPPs. | Shock wave treatment enhances surface area and catalytic activity. |
| Screen-Printed Electrodes (SPEs) | Disposable, portable electrode substrates. | Ideal for field-deployable sensors; ~4 mm diameter working electrode [53]. |
| Phosphate Buffered Saline (PBS) | Electrolyte solution for maintaining stable pH during measurement. | Use at physiological pH for optimal sensor performance [53]. |
| Organophosphate Pesticide Standards | High-purity analytical standards for calibration. | Prepare stock solutions in appropriate solvents (e.g., methanol, acetonitrile). |
| Nafion Perfluorinated Resin | Ion-exchange polymer used as a binder. | Helps form a stable film on the electrode surface. |
Signaling Pathway and Detection Logic
The detection mechanism for electro-inactive OPPs using a metal oxide platform like LaCuO2 is based on a metal-ligand charge transfer process and subsequent signal inhibition, rather than a traditional biochemical pathway.
The increasing use of pesticides in modern agriculture has created a critical need for analytical methods that are not only sensitive and selective but also suitable for on-site, rapid detection. Conventional techniques like high-performance liquid chromatography (HPLC) and gas chromatography-mass spectrometry (GC-MS), while effective, are often costly, time-consuming, and require specialized laboratory settings and trained personnel [11] [3]. This creates a significant gap in our ability to perform routine monitoring and rapid screening. Electrochemical (EC) sensors and optical assays based on surface-enhanced Raman scattering (SERS) have emerged as powerful alternatives, aiming to minimize cost and processing time while improving diagnostic accuracy [54]. Screen-printed electrodes (SPEs) provide an ideal foundation for these sensors due to their portability, low cost, disposability, and ease of mass production [4] [13].
The integration of these two powerful techniques creates electrochemical SERS (EC-SERS), a hybrid electro-optical method that merges the strengths of both approaches [54] [55]. This multimodal approach offers significant advantages: the electrochemical component provides excellent quantitative capabilities and high sensitivity, while SERS contributes unparalleled molecular specificity through its spectral "fingerprint" identification [54]. Furthermore, the combination of SERS detection with other sensing modalities—such as colorimetric, fluorescence, and magnetic detection—creates multimodal biosensors with self-verification capabilities through diverse signal output modalities, effectively improving detection accuracy and reliability [55]. The versatility of nanomaterials offers flexible design solutions for constructing these advanced biosensors, enabling the development of compact, portable, and user-friendly detection systems that can be deployed for on-site monitoring in various settings, from agricultural fields to food processing facilities [55].
Experimental Protocols
Fabrication and Modification of Screen-Printed Electrodes (SPEs)
Objective: To prepare nanomaterial-modified SPEs optimized for EC-SERS applications in pesticide detection.
Materials:
- Commercial carbon-based Screen-Printed Electrodes (e.g., from Metrohm)
- Chloroauric acid (HAuCl₄), silver nitrate (AgNO₃), or other metal precursors
- Raman reporter molecules (e.g., 4-aminothiophenol, 4-nitrothiophenol)
- Functional nanomaterials: Graphene oxide (GO), multi-walled carbon nanotubes (MWCNTs), gold nanoparticles (AuNPs), silver nanoparticles (AgNPs)
- Linking agents: (3-aminopropyl)triethoxysilane (APTES), cysteamine
- Biorecognition elements: Antibodies, aptamers, enzymes (e.g., acetylcholinesterase for organophosphorus pesticides)
- Buffers: Phosphate buffer saline (PBS, 0.1 M, pH 7.4), acetate buffer
Protocol:
Step 1: Electrode Pretreatment
- Clean SPEs electrochemically by performing 10-15 cycles of cyclic voltammetry (CV) in 0.1 M H₂SO₄ from -0.2 to +0.6 V (vs. Ag/AgCl reference) at a scan rate of 50 mV/s.
- Alternatively, treat SPEs with oxygen plasma for 2-5 minutes to introduce surface functional groups and enhance hydrophilicity [4].
Step 2: Nanomaterial Synthesis and Electrode Modification Option A: Graphene Oxide/Carbon Nanotube Modification
- Prepare dispersions of GO (1 mg/mL) and MWCNTs (1 mg/mL) in deionized water using probe sonication for 30 minutes.
- Drop-cast 5-10 μL of the nanomaterial dispersion onto the working electrode surface.
- Allow to dry at room temperature or in an oven at 40°C for 1 hour [4].
Option B: Plasmonic Nanoparticle Decoration
- Synthesize gold nanoparticles (AuNPs) by reducing HAuCl₄ with sodium citrate in boiling water [56].
- Functionalize the nanomaterial-modified SPE with AuNPs by electrodeposition using CV (from -0.8 to +0.8 V for 20 cycles in 1 mM HAuCl₄) or by drop-casting AuNP colloid.
- For SERS enhancement, create "hot spots" by generating nanoparticle aggregates or core-shell structures [56].
Step 3: Immobilization of Recognition Elements
- For aptamer-based sensors: Incubate modified SPEs with 5 μM thiolated aptamer solution in PBS buffer for 12-16 hours at 4°C to form self-assembled monolayers.
- For antibody-based sensors: Activate the surface with EDC/NHS chemistry, then incubate with specific antibodies (10-50 μg/mL) for 2 hours at room temperature.
- For enzyme-based sensors: Immobilize acetylcholinesterase (AChE) by cross-linking with glutaraldehyde vapor for 1 hour [3].
Step 4: Characterization
- Characterize the modified electrode surface using scanning electron microscopy (SEM) to verify nanomaterial distribution.
- Perform electrochemical characterization using CV and electrochemical impedance spectroscopy (EIS) in 5 mM [Fe(CN)₆]³⁻/⁴⁻ to confirm successful modification.
- Assess SERS activity by measuring the Raman signal intensity of a standard reporter molecule.
EC-SERS Detection of Pesticide Residues
Objective: To perform simultaneous electrochemical and SERS detection of target pesticides using modified SPEs.
Materials:
- Modified SPEs from Protocol 2.1
- Target pesticide standards (e.g., parathion, carbaryl, imidacloprid)
- Electrochemical cell or microfluidic chamber
- Raman spectrometer with appropriate laser source (e.g., 785 nm)
- Potentiostat/galvanostat instrument
Protocol:
Step 1: Sample Preparation and Incubation
- Prepare pesticide standards in appropriate buffer or simulate real samples by spiking known concentrations into fruit/vegetable extracts.
- For competitive assays, incubate 50 μL of sample solution with the modified SPE for 15-20 minutes at room temperature with gentle agitation.
- Rinse the electrode gently with buffer to remove unbound molecules.
Step 2: Electrochemical Measurement
- Perform electrochemical measurements in a three-electrode configuration using the modified SPE as working electrode.
- Use differential pulse voltammetry (DPV) with parameters: potential range from -0.2 to +0.6 V, pulse amplitude 50 mV, pulse width 50 ms, step height 10 mV.
- Alternatively, use electrochemical impedance spectroscopy (EIS) with frequency range from 0.1 Hz to 100 kHz at formal potential of the redox probe [54] [3].
- Record the electrochemical signal change relative to pesticide concentration.
Step 3: SERS Measurement
- Place the electrode under the Raman microscope objective (10× or 20×).
- Focus the laser beam (e.g., 785 nm, 10 mW) on the electrode surface.
- Acquire SERS spectra with integration time of 10-30 seconds.
- Monitor characteristic Raman peaks of the pesticide molecules or the signal changes from Raman reporters.
Step 4: Data Analysis
- For electrochemical data: Plot calibration curve of current response vs. pesticide concentration.
- For SERS data: Analyze peak intensity at specific Raman shifts or employ multivariate analysis for fingerprint identification.
- Correlate EC and SERS signals for enhanced detection reliability.
Performance Data and Analysis
The tables below summarize the performance characteristics of various nanomaterial-based sensors for pesticide detection, highlighting the advantages of hybrid approaches.
Table 1: Performance Comparison of Nanomaterial-Based Sensors for Pesticide Detection
| Sensor Type | Nanomaterials Used | Target Pesticide | LOD | Detection Range | Analysis Time | Reference |
|---|---|---|---|---|---|---|
| Enzymatic Electrochemical | CuONPs nanozyme | Malathion | 0.08 mg/L | 0.1-5 mg/L | ~10 min | [3] |
| Fluorescent Microfluidic | CdTe Quantum Dots | Organophosphorus | 0.38 pM | - | - | [3] |
| Colorimetric Aptasensor | Single-atom Ce nanozyme | Organophosphorus | - | - | - | [3] |
| SERS-based | AuNPs/AgNPs | Multiple pesticides | ppt-ppb range | - | < 30 min | [55] |
| EC-SERS Hybrid | AuNP/GO/SPE | Parathion, Carbaryl | sub-ppb | Wide linear range | < 20 min | [54] [55] |
Table 2: Key Performance Advantages of EC-SERS Hybrid Systems
| Parameter | Electrochemical Sensors | SERS Sensors | EC-SERS Hybrid |
|---|---|---|---|
| Sensitivity | Excellent (nM-pM) | Outstanding (single molecule) | Enhanced through dual amplification |
| Selectivity | Moderate to High | High (molecular fingerprint) | Very High (dual verification) |
| Quantitative Ability | Excellent | Moderate | Excellent with internal calibration |
| Multiplexing Capability | Limited | Excellent | Good to Excellent |
| On-site Applicability | Excellent | Moderate to Good | Good with proper design |
| Cost | Low | Moderate | Moderate |
| Sample Throughput | High | Moderate | High with automation |
Visualization Schematics
Workflow for EC-SERS Pesticide Detection
EC-SERS Detection Mechanism
Research Reagent Solutions
Table 3: Essential Research Reagents for EC-SERS Pesticide Detection
| Reagent Category | Specific Examples | Function/Purpose | Key Characteristics |
|---|---|---|---|
| Electrode Materials | Screen-printed carbon electrodes (SPCEs) | Sensor platform/substrate | Portable, cost-effective, disposable [4] |
| Plasmonic Nanomaterials | Gold nanoparticles (AuNPs), Silver nanoparticles (AgNPs) | SERS signal enhancement | Strong plasmonic resonance, tunable morphology [56] |
| Conductive Nanomaterials | Graphene oxide (GO), Carbon nanotubes (CNTs) | Electron transfer enhancement | High surface area, excellent conductivity [4] |
| Recognition Elements | Acetylcholinesterase (AChE), Specific aptamers, Antibodies | Target recognition and binding | High specificity, various affinity options [3] |
| Raman Reporters | 4-aminothiophenol, 4-nitrothiophenol, Methylene blue | SERS signal generation | Large Raman cross-sections, specific fingerprints [56] |
| Electrochemical Probes | Ferricyanide/ferrocyanide, Methylene blue | Redox activity for EC detection | Reversible electrochemistry, well-defined peaks [54] |
The integration of EC-SERS and multimodal detection platforms represents a significant advancement in analytical chemistry, particularly for pesticide analysis. These hybrid systems leverage the complementary strengths of electrochemical and spectroscopic techniques, providing enhanced sensitivity, selectivity, and reliability compared to single-mode detection approaches. The use of nanomaterial-modified screen-printed electrodes as the foundational platform enables the development of portable, cost-effective sensors suitable for field-deployment and point-of-care testing.
Future research should focus on optimizing the synergy between detection modalities, improving the reproducibility of nanomaterial fabrication, developing standardized protocols for sensor calibration and validation, and integrating these systems with microfluidics and data analysis algorithms for fully automated operation. As these technologies mature, EC-SERS hybrid systems hold tremendous potential to transform environmental monitoring, food safety assurance, and public health protection through rapid, accurate, and on-site detection of pesticide residues and other contaminants.
Performance Enhancement and Challenge Mitigation in Real-World Applications
The performance of electrochemical sensors, particularly for critical applications like pesticide analysis, is profoundly influenced by the properties of the nanomaterials used to modify the electrode surfaces. For researchers developing nanomaterial-modified screen-printed electrodes (SPEs), a cornerstone of modern electroanalysis, understanding the synthesis-structure-property relationships is essential [57]. The intentional engineering of nanomaterial size, morphology, and composition directly controls key sensor performance metrics, including sensitivity, limit of detection (LOD), and selectivity [58] [11]. This Application Note details the quantitative impacts of these parameters and provides standardized protocols for leveraging these relationships to develop advanced sensors for pesticide detection.
Quantitative Impact of Nanomaterial Properties on Sensor Performance
The integration of nanomaterials into electrochemical sensors significantly enhances performance by increasing the electroactive surface area, improving electron transfer kinetics, and providing catalytic activity [58] [59]. The following tables summarize the specific effects of nanomaterial properties on sensor performance, with a focus on pesticide detection.
Table 1: Impact of Nanomaterial Composition and Morphology on Sensor Performance for Pesticide Detection
| Nanomaterial Composition & Morphology | Key Properties Enhanced | Target Pesticide(s) | Reported LoD | Ref. |
|---|---|---|---|---|
| Carbon Nanotubes (CNTs) (1D cylindrical nanostructure) | High surface area, excellent electrical conductivity, p-type semiconducting behavior | NO₂, NH₃ | ppm-level | [60] |
| Gold Nanoparticles (AuNPs) (Spherical) | High conductivity, catalytic activity, biocompatibility, facile functionalization | Organophosphorus, Methomyl, Chlorpyrifos | 19-81 ng L⁻¹ | [32] |
| Silver Nanoparticles (AgNPs) (Spherical) | Unique optical & electrical properties, high surface-to-volume ratio | Information Missing | Information Missing | [32] |
| Metal-Organic Frameworks (MOFs) (Porous crystalline) | Ultra-high surface area, tunable porosity, selective adsorption | Methyl Parathion | Ultrasensitive detection | [11] |
| Binary Nanocomposite (e.g., Metal NP/CNT) | Synergistic effects, superior properties beyond individual components | Various | Enhanced sensitivity and lower LOD | [58] |
Table 2: Influence of Nanomaterial Size and Electrode Modification on Analytical Performance
| Parameter | Influence on Sensor Properties | Effect on Sensor Performance |
|---|---|---|
| Particle Size | • Increased surface-to-volume ratio • Altered electronic structure (quantum effects) • Increased catalytic activity | • Higher sensitivity • Lower Limit of Detection (LOD) • Improved electrocatalytic response |
| Electrode Effective Surface Area | • Increased number of active sites for electron transfer • Enhanced analyte adsorption | • Stronger electrochemical signal • Improved signal-to-noise ratio |
| Use of Nanocomposites | • Combination of advantageous properties (e.g., conductivity + catalysis) • Prevention of nanomaterial aggregation | • Wider linear detection range • Enhanced stability and reproducibility • Selective analyte recognition |
Experimental Protocols for Sensor Fabrication and Evaluation
Protocol: Dielectrophoretic (DEP) Trapping of Nanomaterials on Microelectrodes
This bottom-up approach allows for the precise assembly and simultaneous electrical characterization of nanomaterials like CNTs or nanowires between microelectrodes [60].
- Principle: Dielectrophoresis (DEP) is the motion of dielectrically polarized particles in a non-uniform electric field. Positive DEP attracts nanomaterials toward regions of high electric field strength, such as electrode gaps [60].
- Materials:
- Interdigitated metallic microelectrode array (e.g., Cr or Au with 5 µm gap).
- Aqueous suspension of nanomaterials (e.g., MWCNTs, ZnO nanowires).
- Function generator.
- Impedance analyzer.
- Microfluidic chamber or silicon rubber spacer.
- Step-by-Step Procedure:
- Suspension Preparation: Disperse the nanomaterial powder in an aqueous medium (e.g., deionized water) using ultrasonication. Remove large aggregates by centrifugation or filtration.
- Setup Configuration: Place a droplet of the nanomaterial suspension into the microelectrode chamber. Alternatively, continuously pump the suspension through a microfluidic cell mounted over the electrode.
- DEP Trapping: Apply an AC voltage (e.g., 10 Vpp, 100 kHz for CNTs) across the microelectrodes using a function generator. Monitor the electrode impedance in real-time using an impedance analyzer.
- Termination and Drying: Once the impedance signal indicates a sufficient amount of nanomaterial has been trapped (forming bridges across the electrode gap), stop the AC voltage. Gently evaporate the aqueous medium at room temperature to immobilize the nanostructures on the electrode via van der Waals forces.
- Notes: The DEP force is highly dependent on the AC field frequency, which can be tuned to selectively trap nanomaterials based on their dielectric properties (e.g., metallic vs. semiconducting CNTs) [60]. Real-time impedance monitoring (DEPIM) allows for precise control and calibration of the sensor's initial conductance.
Protocol: Modifying Screen-Printed Electrodes (SPEs) with Nanomaterial Inks
Drop-casting is a widely used, straightforward method for modifying commercial SPEs with functional nanomaterials.
- Principle: A stable dispersion (ink) of nanomaterials is applied directly to the working electrode surface. The solvent evaporates, leaving a thin film of the nanomaterial that enhances the electrode's properties [58] [11].
- Materials:
- Commercial or in-house fabricated SPEs.
- Nanomaterial (e.g., graphene oxide, MWCNTs, AuNPs).
- Dispersion solvent (e.g., ethanol, DMF, water).
- Binder/Stabilizer (e.g., Nafion, chitosan).
- Precision micropipette.
- Step-by-Step Procedure:
- Ink Preparation: Prepare a homogeneous ink by dispersing the nanomaterial in a suitable solvent (e.g., 1 mg/mL in ethanol). Sonication for 30-60 minutes is typically required. To enhance stability and film formation, add a small amount of binder (e.g., 0.5% v/v Nafion).
- Electrode Pretreatment: Clean the SPE's working electrode surface according to manufacturer instructions, if applicable (e.g., electrochemical cycling in a blank solution).
- Modification: Using a precision micropipette, deposit a small, controlled volume (e.g., 2-10 µL) of the nanomaterial ink onto the working electrode.
- Drying: Allow the modified electrode to dry under ambient conditions or under an infrared lamp. The formation of a uniform film is critical for reproducible results.
- Notes: The performance of the modified electrode is highly reproducible if the concentration of the ink and the volume deposited are kept consistent. The use of binders like Nafion can also impart selectivity by forming a charge-selective membrane [58].
Workflow and Property-Performance Relationships
The following diagram illustrates the logical and experimental workflow for developing and optimizing a nanomaterial-based electrochemical sensor, from material selection to performance validation.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for Nanomaterial-Based Sensor Development
| Item | Function/Application | Key Characteristics |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Miniaturized, disposable, portable platform for electrochemical analysis. | Cost-effective, mass-producible, ideal for field-deployable sensors [59] [11]. |
| Carbon Nanotubes (CNTs) | Electrode modifier for enhancing electron transfer and surface area. | High conductivity, large specific surface area, (electro)chemical stability [58] [60]. |
| Metal Nanoparticles (Au, Ag) | Electrode modifier for catalytic signal amplification. | High conductivity, biocompatibility, tunable optical & catalytic properties [32]. |
| Nafion | Cation-exchange polymer used as a binder and selective membrane. | Prevents fouling, imparts selectivity, stabilizes nanomaterial film on electrode [58]. |
| Chitosan | Natural biopolymer used as a dispersing agent and immobilization matrix. | Biocompatibility, biodegradability, non-toxicity, excellent film-forming ability [11]. |
| UV-Curable Polymers (e.g., NOA 81) | Substrate for nano-imprint lithography of plasmonic sensors. | Enables high-resolution, low-cost fabrication of nanostructured sensor surfaces [61]. |
Electrochemical sensors employing nanomaterial-modified screen-printed electrodes (SPEs) present powerful tools for pesticide analysis, offering portability, cost-effectiveness, and high sensitivity [62] [63]. However, their analytical performance and operational lifetime are severely compromised by electrode fouling and instability, particularly when dealing with complex environmental and agricultural samples [64] [63]. Fouling occurs through the non-specific adsorption of matrix components—such as proteins, organic matter, and pesticide metabolites—onto the electrode surface, leading to passivation of active sites, reduced electron transfer kinetics, signal drift, and ultimately, analytical failure [65] [66]. These challenges are pronounced in the context of a research thesis focused on pesticide analysis, where samples often include soil extracts, plant tissues, and water with significant organic content [63] [66]. Within this thesis framework, which investigates nanomaterial-modified SPEs for pesticide detection, developing robust strategies to mitigate fouling is not merely an optimization step but a fundamental requirement for generating reliable, reproducible data. This document provides detailed application notes and protocols for implementing effective surface passivation and regeneration strategies, serving as an essential toolkit for enhancing the rigor and validity of electrochemical research.
Surface Passivation Strategies
Surface passivation involves the application of a protective layer or modification to the electrode surface to shield it from fouling agents while maintaining, or even enhancing, its electrochemical activity towards the target analyte.
Nanomaterial-Based Passivation Layers
Incorporating specific nanomaterials can create selective barriers that minimize non-specific adsorption.
- Carbon Nanomaterial Coatings: Graphene oxide (GO) possesses residual oxygen-containing functional groups that can improve selectivity for different targets and favor the diffusion of probes. A protocol for modifying SPEs with GO is provided in Section 4.1 [65].
- Metallic Nanoparticle and Rock Salt Layers: Gold nanoparticles (AuNPs) can be synthesized and deposited to enhance conductivity and provide a stable, biocompatible surface that resists fouling [67]. Furthermore, a strategy of surface reduction passivation can be adapted from battery research. This involves forming a thin rock salt passivation layer on the surface of materials to suppress interfacial side reactions, lattice oxygen loss, and phase transition, thereby greatly improving interface stability [68].
Chemical and Biomolecular Passivation Layers
- Self-Assembled Monolayers (SAMs): On gold SPEs, alkanethiols like 11-mercaptoundecanoic acid (MUA) can form dense, ordered monolayers that prevent non-specific adsorption through steric hindrance and surface energy modification [65].
- Polymer Coatings: Membranes of polymers like Nafion or cellulose nanocrystals (CNCs) can be cast on the electrode surface. CNCs, for instance, form a robust, hydrophilic network that acts as a physical barrier to larger interferents [67].
- Protein Blocking: A common biosensing strategy involves using inert proteins like Bovine Serum Albumin (BSA) to block residual active sites on the modified electrode surface after probe immobilization, thereby reducing non-specific binding [65].
Table 1: Summary of Surface Passivation Strategies for Fouling Mitigation
| Strategy | Key Components | Mechanism of Action | Best Suited For |
|---|---|---|---|
| Nanomaterial Coatings | Graphene Oxide, Carbon Nanotubes, AuNPs | Creates a selective barrier; enhances electron transfer; minimizes adsorption [62] [67] [65]. | Broad-spectrum pesticide detection in complex matrices. |
| Chemical Membranes | Cellulose Nanocrystals, Nafion | Forms a physical, hydrophilic/hydrophobic barrier that excludes interferents based on size and charge [67]. | Analysis in turbid water or soil extracts. |
| Molecular Monolayers | 11-Mercaptoundecanoic Acid | Forms a dense, ordered layer on gold surfaces, preventing fouling via steric hindrance [65]. | Gold-based SPEs used in biosensor configurations. |
| Surface Reduction Passivation | H2/Ar atmosphere | Forms a stabilizing rock salt layer to suppress surface reactions and degradation [68]. | High-stress analysis requiring long-term electrode stability. |
The following diagram illustrates the decision-making workflow for selecting and implementing an appropriate passivation strategy within an experimental design.
Surface Regeneration and Cleaning Protocols
When passivation is insufficient, or for reusable electrodes, regeneration of the fouled surface is necessary. The following protocols detail effective cleaning methods.
Electrochemical Cleaning of Screen-Printed Gold Electrodes (SPGEs)
This protocol, adapted from recent research, effectively removes fouling layers and restores the electrochemical activity of SPGEs [65].
Principle: Application of cyclical potentials in a cleaning solution to oxidize and reduce adsorbed organic contaminants, followed by electrochemical characterization to confirm surface cleanliness [65].
Reagents:
- Cleaning Solution: 3% v/v H2O2 in 0.1 M HClO4 (Perchloric Acid).
- Characterization Solution: 2.5 mM Potassium Ferricyanide/Ferrocyanide ([Fe(CN)6]3−/4−) in 0.01 M PBS, pH 7.4.
- Milli-Q water.
Procedure:
- Initial Characterization: Perform a Cyclic Voltammetry (CV) scan on the fouled SPGE in the characterization solution (parameters below) to record the degraded state.
- Cleaning: Apply 150 µL of the cleaning solution to the SPGE.
- Run 10 cycles of CV with the following parameters:
- Potential Range: -700 mV to +2000 mV (vs. Ag/AgCl reference).
- Scan Rate: 100 mV/s.
- Rinsing: Thoroughly rinse the electrode with Milli-Q water to remove all traces of the cleaning solution.
- Activation/Stabilization: Perform 10 additional CV cycles in a fresh aliquot of the characterization solution using a standard CV parameters (e.g., -400 mV to +500 mV, 50 mV/s) to stabilize the electrode surface.
- Final Characterization: Record a new CV in the characterization solution. A restored, well-defined redox peak for the [Fe(CN)6]3−/4− couple indicates successful regeneration [65].
Validation: Compare the peak currents and peak-to-peak separation (ΔEp) before and after cleaning. A significant increase in current and a decrease in ΔEp confirm improved electron transfer and successful de-fouling [65].
Chemical Incubation Cleaning
A simpler, chemical-only method can also be effective for certain types of fouling [65].
Procedure:
- Apply 150 µL of the cleaning solution (3% H2O2 and 0.1 M HClO4) to the SPGE.
- Allow it to incubate at rest for 10 minutes.
- Rinse the electrode thoroughly with Milli-Q water.
- Perform electrochemical characterization as described above to validate effectiveness.
Table 2: Electrode Regeneration Protocols: A Comparative Analysis
| Parameter | Electrochemical Cleaning | Chemical Incubation |
|---|---|---|
| Principle | Electrochemical oxidation/reduction of contaminants [65]. | Chemical oxidation of organic foulants [65]. |
| Key Reagents | H2O2/HClO4 solution; [Fe(CN)6]3−/4− for validation [65]. | H2O2/HClO4 solution [65]. |
| Procedure Complexity | Moderate (requires potentiostat) [65]. | Simple (incubation only) [65]. |
| Typical Efficacy | High (effectively removes most organic layers) [65]. | Moderate (may be insufficient for tenacious films) [65]. |
| Risk of Surface Damage | Moderate (controlled by potential window and cycle number) [65]. | Lower (gentler on electrode materials) [65]. |
| Validated For | Screen-printed gold electrodes (SPGEs) [65]. | Screen-printed gold electrodes (SPGEs) [65]. |
Detailed Experimental Protocols for Key Setups
Protocol: Modification of SPE with Graphene Oxide for Enhanced Selectivity
This protocol details the preparation of a graphene oxide-modified screen-printed electrode, which can exhibit improved resistance to fouling and enhanced selectivity for certain analytes [65].
Reagents:
- Graphene Oxide (GO) dispersion (e.g., 1 mg/mL in water).
- Phosphate Buffered Saline (PBS), pH 7.4.
- Screen-printed carbon electrodes (SPCEs).
- Ethanol and Milli-Q water.
Procedure:
- SPCE Pre-treatment: If required, pre-treat the SPCE via plasma treatment or electrochemical activation to introduce surface functional groups.
- GO Dispersion Preparation: Dilute the commercial GO stock to a concentration of 0.5 mg/mL and sonicate for 30 minutes to ensure a homogeneous dispersion.
- Drop-Casting: Pipette a precise volume (e.g., 5-10 µL) of the GO dispersion directly onto the working electrode surface of the SPCE.
- Drying: Allow the electrode to dry under ambient conditions or in a desiccator until all solvent has evaporated, leaving a thin GO film.
- Rinsing: Gently rinse the modified electrode (GO/SPCE) with PBS to remove any loosely adsorbed GO sheets.
- Characterization: Characterize the modified electrode using Cyclic Voltammetry (CV) and Electrochemical Impedance Spectroscopy (EIS) in a standard redox probe like [Fe(CN)6]3−/4− to confirm successful modification and improved surface properties [65].
Protocol: Assembling a Nanomaterial-Modelled Electrode for Pesticide Detection
This general protocol outlines the steps for constructing a sensor for organophosphate pesticide analysis, integrating passivation strategies.
Reagents:
- Acetylcholinesterase (AChE) enzyme.
- Gold Nanoparticles (AuNPs) solution.
- Chitosan solution (0.5% w/v in acetic acid).
- Screen-printed electrode (Gold or Carbon).
- Substrate and standard solutions for the target pesticide.
Procedure:
- Electrode Modification: a. If using a carbon SPE, deposit AuNPs via drop-casting or electrodeposition to enhance conductivity and provide a platform for biomolecule immobilization. b. Apply a passivation layer. For instance, cast a thin layer of chitosan, which acts as both a biocompatible matrix and a mild fouling-resistant layer.
- Biosensor Assembly: a. Immobilize the AChE enzyme on the modified electrode by physically adsorbing it onto the chitosan/AuNP/SPE or through cross-linking. b. Incubate and then rinse to remove unbound enzyme.
- Inhibition Assay for Pesticide Detection: a. Incubate the biosensor with a sample containing the target organophosphate pesticide for a fixed time (e.g., 10-15 minutes). The pesticide will inhibit the AChE enzyme. b. Wash the electrode to stop the inhibition reaction. c. Transfer the electrode to a standard electrochemical cell containing a known concentration of the enzyme substrate (e.g., acetylthiocholine). d. Measure the amperometric current generated by the enzymatic product (thiocholine). The degree of signal reduction is proportional to the pesticide concentration [63].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents for Electrode Passivation and Regeneration
| Reagent / Material | Function / Application | Notes & Considerations |
|---|---|---|
| Graphene Oxide | Nanomaterial coating for SPEs; enhances selectivity and can resist fouling [65]. | Concentration and dispersion quality are critical for reproducible film formation. |
| Gold Nanoparticles | Enhances conductivity and active surface area; platform for SAMs and biomolecule attachment [67]. | Synthesized via chemical reduction (e.g., citrate method); size must be controlled. |
| Cellulose Nanocrystals | Biocompatible polymer coating; forms a physical barrier against interferents [67]. | Provides a hydrophilic surface that resists protein adsorption. |
| 11-Mercaptoundecanoic Acid | Forms a self-assembled monolayer on gold surfaces to prevent non-specific adsorption [65]. | Requires pure ethanol as a solvent and several hours for a well-ordered monolayer to form. |
| H2O2 / HClO4 Solution | Electrochemical cleaning solution for regenerating gold SPGEs [65]. | Caution: Perchloric acid is a strong oxidizer and requires careful handling. |
| Potassium Ferricyanide | Redox probe for electrode characterization via CV and EIS [65]. | A well-defined CV signal indicates a clean, active surface. A degraded signal suggests fouling. |
| Bovine Serum Albumin | Blocking agent to passivate unused active sites on biosensor surfaces [65]. | Typically used as a 1% w/v solution in buffer; incubate for 30-60 minutes. |
In analytical chemistry, particularly in the detection of pesticides within complex matrices using advanced electrochemical sensors, matrix interference presents a significant challenge that can compromise data accuracy and reliability. Matrix effects (ME) are defined as the combined influence of all sample components other than the analyte on the measurement of quantity [69]. In the specific context of a broader thesis on nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, minimizing these effects is paramount to developing robust, sensitive, and field-deployable analytical methods [11] [10]. Nanomaterial-modified SPEs offer advantages such as portability, low cost, and high sensitivity [10]. However, when deployed for analysis in real-world samples like food, environmental water, or biological fluids, the electrodes encounter a multitude of interfering substances, including proteins, phospholipids, salts, and organic acids, which can alter the electrochemical response [69].
The core of the problem lies in the fact that these interferents can co-elute or coexist with the target pesticide analytes, leading to phenomena such as ion suppression or enhancement in mass spectrometry [69] [70], or fouling and signal suppression in electrochemical sensors [11]. For electrochemical (bio)sensors, this can manifest as blocked active sites on the nanomaterial surface, reduced charge transfer efficiency, or non-specific binding, ultimately affecting parameters like limits of detection, reproducibility, and accuracy [10]. Therefore, effective sample preparation and preconcentration are not merely preliminary steps but are critical components in the workflow to ensure the analytical validity of data generated by nanomaterial-modified SPEs.
Understanding and Assessing Matrix Effects
Before developing strategies for minimization, it is essential to properly identify and evaluate the presence and extent of matrix effects. Several established methods can be employed, each providing different levels of qualitative or quantitative insight.
Post-Column Infusion Method
This technique offers a qualitative assessment of matrix effects throughout the chromatographic run, identifying regions of ion suppression or enhancement [69] [70]. The analysis is performed by injecting a blank sample extract into the LC system while a solution of the analyte is infused post-column via a T-piece. A stable signal indicates no matrix effects, whereas a dip or rise in the baseline at specific retention times indicates ion suppression or enhancement, respectively [69]. This method is particularly useful in the early stages of method development to pinpoint problematic retention time windows.
Post-Extraction Spike Method
This method provides a quantitative measure of matrix effects by comparing the analytical response of an analyte in a pure solvent to its response when spiked into a blank matrix sample that has already undergone the sample preparation process [69] [70]. The Matrix Effect (ME%) is calculated as follows:
ME% = (B / A) × 100%
Where A is the peak response of the analyte in neat solvent and B is the peak response of the analyte spiked into the processed blank matrix. An ME% of 100% indicates no matrix effect, <100% indicates ion suppression, and >100% indicates ion enhancement [70].
Slope Ratio Analysis
A semi-quantitative extension of the post-extraction spike method, this approach involves creating calibration curves using both solvent-based standards and matrix-matched standards spiked post-extraction across a range of concentrations [69]. The ratio of the slopes of these two calibration curves provides an average measure of the matrix effect across the entire calibration range.
Table 1: Comparison of Matrix Effect Assessment Methods
| Method Name | Type of Data | Description | Key Limitations |
|---|---|---|---|
| Post-Column Infusion [69] [70] | Qualitative | Identifies retention time zones with ion suppression/enhancement via constant analyte infusion. | Does not provide quantitative data; requires specialized setup. |
| Post-Extraction Spike [69] [70] | Quantitative | Compares analyte response in solvent vs. spiked blank matrix at a single concentration. | Requires availability of a blank matrix. |
| Slope Ratio Analysis [69] | Semi-Quantitative | Compares slopes of calibration curves in solvent vs. matrix across a concentration range. | Provides an average value, may mask concentration-dependent effects. |
The following workflow (Figure 1) outlines the logical process for a researcher to assess and then mitigate matrix effects in their analytical method.
Figure 1: Decision Workflow for Managing Matrix Effects. This diagram outlines the systematic process for assessing, minimizing, and, if necessary, compensating for matrix effects during analytical method development.
Protocols for Sample Preparation and Preconcentration
Effective sample preparation is the first line of defense against matrix interference. The goal is to isolate the analyte from the complex matrix and, if possible, concentrate it to improve detection sensitivity.
Solid-Phase Extraction (SPE) for Complex Matrices
SPE is a widely used technique for cleaning up samples and preconcentrating analytes.
Detailed Protocol:
- Conditioning: Sequentially pass 5-10 mL of methanol and 5-10 mL of water or a buffer compatible with the sorbent through the SPE cartridge. Do not allow the sorbent to dry out.
- Loading: Adjust the sample pH if necessary to ensure the target pesticides are in a neutral form for reversed-phase SPE. Load the sample (e.g., 100-500 mL of environmental water) through the cartridge at a controlled flow rate of 2-5 mL/min.
- Washing: Remove weakly retained matrix interferents by passing 5-10 mL of a wash solution (e.g., 5-20% methanol in water). This step is crucial for eliminating salts and polar organic compounds.
- Elution: Elute the target pesticides with 5-10 mL of a strong organic solvent (e.g., acetonitrile, methanol, or a mixture with dichloromethane for non-polar pesticides). Collect the eluate in a clean tube.
- Reconstitution: Evaporate the eluate to dryness under a gentle stream of nitrogen. Reconstitute the dry residue in 0.5-1.0 mL of the initial mobile phase or a solvent compatible with the subsequent analysis on the nanomaterial-modified SPE.
Protein Precipitation for Biological Fluids
This is a simple and rapid technique for removing proteins from samples like plasma or serum.
Detailed Protocol:
- Precipitation: Mix a 100 µL aliquot of the biological sample with 300 µL of a precipitant solvent (e.g., acetonitrile or methanol) in a 1.5 mL microcentrifuge tube. Vortex vigorously for 1-2 minutes.
- Centrifugation: Centrifuge the mixture at 10,000-15,000 × g for 10 minutes to pellet the precipitated proteins.
- Collection: Carefully transfer the clear supernatant to a new clean tube.
- Evaporation and Reconstitution (Optional): Evaporate the supernatant to dryness under nitrogen gas and reconstitute in a suitable buffer for electrochemical analysis. This step also serves as a preconcentration method.
Dilution to Minimize Matrix Effects
In cases where the analytical method is sufficiently sensitive, simple dilution of the sample with a compatible solvent can be an effective way to reduce the concentration of interfering substances [70].
Detailed Protocol:
- Perform a preliminary experiment to determine the extent of matrix effects using the post-extraction spike method.
- If matrix effects are observed, prepare a series of sample dilutions (e.g., 1:2, 1:5, 1:10) with a solvent like water or mobile phase buffer.
- Re-analyze the diluted samples and re-assess the matrix effects. Select the dilution factor that adequately reduces the matrix effect while maintaining the analyte concentration above the limit of quantification of the sensor.
Advanced Mitigation Strategies and Calibration Techniques
When sample preparation alone is insufficient to fully overcome matrix effects, advanced strategies and specific calibration techniques must be employed.
Chromatographic Optimization
Adjusting chromatographic parameters can spatially separate analytes from interferents, a strategy that can be analogously applied to the "separation" step in an electrochemical sensor's cleaning or conditioning protocol.
- Adjusting Mobile Phase and Gradient: Modifying the pH, buffer concentration, or organic solvent strength can shift the retention times of analytes and interferents, preventing their co-elution onto the sensor surface [69] [70].
- Column Chemistry Selection: Using alternative column chemistries (e.g., HILIC instead of reversed-phase) can provide a different selectivity, potentially resolving the analyte from matrix components [70].
Calibration Techniques to Compensate for Residual Effects
These techniques are essential for achieving accurate quantification when residual matrix effects persist.
- Matrix-Matched Calibration: Calibration standards are prepared in a blank matrix that is free of the analyte. This calibrates the system response to mimic that of the real samples [69] [70]. The primary challenge is sourcing a truly blank matrix, which is impossible for endogenous analytes.
- Standard Addition: This method is highly effective when a blank matrix is unavailable [70]. The sample is divided into several aliquots, and increasing known amounts of the analyte standard are spiked into them. The analytical response is measured for each aliquot, and the original concentration is determined by extrapolating the calibration curve back to the x-axis.
- Internal Standardization: This is the gold standard for compensating for both matrix effects and instrument variability.
- Stable Isotope-Labeled Internal Standard (SIL-IS): An isotopically labeled version of the analyte (e.g., deuterated) is the ideal choice as it has nearly identical chemical properties and co-elutes with the analyte, perfectly compensating for matrix effects [69] [70].
- Structural Analogue Internal Standard: If a SIL-IS is unavailable or too costly, a structurally similar compound that behaves similarly in the analytical system can be used as a cheaper alternative [70].
Table 2: Summary of Mitigation and Compensation Strategies for Matrix Effects
| Strategy Category | Technique | Principle | Advantages | Disadvantages | |
|---|---|---|---|---|---|
| Sample Preparation | Solid-Phase Extraction (SPE) | Selective retention of analyte or interferents on a sorbent. | Effective cleanup and preconcentration. | Can be time-consuming; method development needed. | |
| Protein Precipitation | Denaturation and removal of proteins. | Simple, fast, and high recovery. | Less selective; may not remove all interferents. | ||
| Sample Dilution [70] | Reduces concentration of interferents. | Extremely simple and fast. | Requires high analytical sensitivity. | ||
| Analytical Separation | Chromatographic Optimization [69] [70] | Alters retention to separate analyte from interferents. | Can be highly effective without extra steps. | Time-consuming optimization; may not work for all co-elutions. | |
| Calibration | Matrix-Matched Calibration [69] [70] | Standards prepared in blank matrix. | Conceptually simple and effective. | Blank matrix not always available. | |
| Standard Addition [70] | Analyte is spiked at different levels into the sample itself. | Does not require a blank matrix. | Labor-intensive for large sample batches. | ||
| Internal Standard (IS) | SIL-IS [69] [70] | Isotope-labeled version of the analyte. | Excellent compensation; corrects for losses. | Expensive; not always commercially available. | |
| Structural Analogue IS [70] | Chemically similar compound to the analyte. | More affordable than SIL-IS. | May not perfectly mimic analyte behavior. |
The Scientist's Toolkit: Essential Reagents and Materials
The following table details key reagents, materials, and instruments essential for implementing the protocols described in this application note.
Table 3: Research Reagent Solutions and Essential Materials
| Item Name | Function / Application | Specific Examples / Notes |
|---|---|---|
| Solid-Phase Extraction (SPE) Cartridges | Sample clean-up and preconcentration by retaining analytes or interferents. | Reversed-phase C18 for non-polar pesticides; Mixed-mode cation/anion exchange for ionic compounds. |
| Internal Standards | Compensation for matrix effects and analyte loss during preparation. | Stable Isotope-Labeled Internal Standards (SIL-IS, e.g., Creatinine-d3); Structural Analogues (e.g., Cimetidine) [70]. |
| Nanomaterial-Modified Screen-Printed Electrodes (SPEs) | The sensing platform for electrochemical detection of pesticides. | Working electrode can be modified with carbon nanotubes, graphene, metal nanoparticles (Au, Pt), or metal-organic frameworks (MOFs) to enhance sensitivity and selectivity [11] [10]. |
| Protein Precipitants | Removal of proteins from biological samples. | Acetonitrile, Methanol, Trichloroacetic acid. |
| HPLC-MS Grade Solvents | Used for mobile phase preparation, sample reconstitution, and extraction to minimize background noise. | Acetonitrile, Methanol, Water (with 0.1% formic acid or ammonium acetate) [70]. |
| LC-MS/MS System | High-sensitivity separation and detection of analytes; used for method development and validation. | Used with API sources like Electrospray Ionization (ESI) or Atmospheric Pressure Chemical Ionization (APCI) [69]. |
| Portable Potentiostat | Enables field-deployment and point-of-care use of electrochemical sensors. | Compact instrument for performing voltammetry (CV, DPV, SWV) and impedance spectroscopy (EIS) with SPEs [10]. |
In the field of electrochemical analysis for pesticides, selectivity is the cornerstone of reliability. It ensures a sensor responds exclusively to the target analyte amidst a complex soup of chemical interferents commonly found in environmental and food samples. Cross-reactivity occurs when a sensor's recognition element interacts with non-target molecules that share structural similarities with the analyte, leading to false positives or an overestimation of concentration. For nanomaterial-modified screen-printed carbon electrodes (SPCEs), which are prized for their portability and suitability for on-site analysis, achieving high selectivity is both a critical challenge and a primary research focus [11] [10].
The strategic integration of advanced recognition elements is the most effective pathway to mitigate cross-reactivity. These elements are biological or biomimetic molecules engineered for a highly specific, lock-and-key interaction with a single pesticide or a defined class. By moving beyond simple, non-specific chemical interactions and leveraging the unique properties of nanomaterials, researchers can design sensor surfaces that dramatically reject interferents. This document details the application and protocols for using these advanced elements—namely, antibodies, aptamers, enzymes, and molecularly imprinted polymers (MIPs)—to engineer selectivity into SPCE-based pesticide sensors [3] [71].
Advanced Recognition Elements: Mechanisms and Applications
The selection of a recognition element dictates the fundamental mechanism of detection and the subsequent strategy for minimizing cross-reactivity. The following sections explore the four primary categories of elements used in advanced pesticide sensors.
Antibodies and Immunosensors
Antibodies are Y-shaped proteins produced by the immune system, functioning as highly specific biological recognition elements in immunosensors. The specificity originates from the variable regions at the tips of the antibody's "Y" structure, which recognize and bind to a specific molecular structure, or epitope, on the target pesticide (antigen) [71]. This interaction is driven by a combination of hydrogen bonding, van der Waals forces, and electrostatic interactions [71]. To detect small molecule pesticides, they are typically conjugated to a larger carrier protein to form an immunogenic hapten-protein complex for antibody production [71].
Immunosensors are classified as either labeled or label-free. Labeled immunosensors use tags (e.g., enzymes, metal nanoparticles) attached to the antibody or antigen to generate a measurable electrochemical signal after the binding event, often providing higher sensitivity. In contrast, label-free immunosensors directly measure changes in electrical properties (e.g., capacitance, charge transfer resistance) upon antigen binding, offering simplicity but sometimes at the cost of lower sensitivity [71].
- Cross-Reactivity Reduction Mechanism: The high affinity and specificity of the antibody-antigen interaction are the first line of defense against cross-reactivity. The strength of this interaction is characterized by the affinity constant, and high-affinity antibodies are less likely to bind to structurally similar but non-target compounds. Furthermore, the use of monoclonal antibodies, which are identical and bind to a single epitope, offers superior specificity compared to polyclonal antibodies, which are a mixture targeting multiple epitopes [71].
- Typical Performance Metrics: Immunosensors often achieve detection limits in the picomolar (pM) to nanomolar (nM) range, making them among the most sensitive biosensing platforms [71].
Aptamers and Aptasensors
Aptamers are short, single-stranded DNA or RNA oligonucleotides engineered through an in vitro process (SELEX) to bind with high affinity to a specific target molecule, from small pesticides to large proteins [10] [3]. They are often called "chemical antibodies" but offer several advantages, including superior stability, easier and cheaper production, and the ability to be chemically synthesized and modified [3].
In aptasensors, the aptamer is immobilized on the SPCE surface. Upon binding the target pesticide, the aptamer may undergo a conformational change (e.g., from a random coil to a G-quadruplex), which alters the electrochemical properties at the electrode interface, enabling detection [10].
- Cross-Reactivity Reduction Mechanism: The three-dimensional binding pocket formed by the folded aptamer is highly specific to the size, shape, and functional groups of its target. This precise spatial compatibility allows it to discriminate between closely related chemical analogues. Their synthetic origin also allows for precise sequence control to minimize off-target binding.
- Typical Performance Metrics: Aptasensors are capable of high sensitivity, with some reports achieving detection limits comparable to immunosensors, down to the pM level [72].
Enzymes and Enzymatic Biosensors
Enzymatic biosensors primarily operate through two mechanisms: enzymatic inhibition or catalytic hydrolysis [10]. Acetylcholinesterase (AChE) is the most common enzyme used for organophosphate (OP) and carbamate pesticide detection. In a typical inhibition assay, the enzyme's activity is measured by its catalysis of a substrate (e.g., acetylthiocholine), producing an electroactive product (e.g., thiocholine). The presence of the pesticide inhibits AChE, reducing the product formation and thus the electrochemical signal [10] [3].
- Cross-Reactivity Reduction Mechanism: Enzymatic sensors are inherently less specific than immunosensors or aptasensors, as they detect any compound that inhibits the enzyme. Therefore, their primary strength is class-selective detection (e.g., for all OPs) rather than specific compound identification. Specificity can be improved by using enzymes with narrow substrate specificity or by coupling with chromatographic separation. The emergence of nanoenzymes (nanomaterials with enzyme-mimicking activity) and single-atom nanozymes (SAzymes) offers enhanced stability and can be engineered for improved selectivity [3].
- Typical Performance Metrics: These sensors are highly sensitive to the class of inhibitors, often achieving detection in the nM range, but cannot distinguish between different pesticides within the same class without additional strategies [10].
Molecularly Imprinted Polymers (MIPs)
MIPs are synthetic polymer networks containing tailor-made cavities that function as artificial receptors [3]. They are created by polymerizing functional monomers around a template molecule (the target pesticide). After polymerization, the template is removed, leaving behind cavities that are complementary in size, shape, and functional group orientation to the target.
- Cross-Reactivity Reduction Mechanism: The "memory" effect of the MIP cavity provides excellent molecular recognition. The selectivity arises from the specific spatial arrangement of functional groups within the cavity that form non-covalent bonds (e.g., hydrogen bonds, van der Waals forces) only with the intended target molecule. This makes MIPs highly effective at rejecting molecules of different sizes or with different functional groups.
- Typical Performance Metrics: MIP-based sensors are valued for their high physical and chemical stability, low cost, and reusability. Their sensitivity is continually improving with the integration of nanomaterials, with many sensors reaching nM detection limits [3].
Table 1: Comparison of Advanced Recognition Elements for Cross-Reactivity Reduction
| Recognition Element | Mechanism of Action | Key Advantage for Selectivity | Primary Limitation | Common Pesticide Targets |
|---|---|---|---|---|
| Antibodies | High-affinity binding to a specific antigenic epitope | Very high specificity; can generate monoclonal antibodies for a single compound | Susceptible to denaturation; production can be complex and costly | Organophosphates, Triazines, Neonicotinoids [71] |
| Aptamers | Target-induced folding/ conformational change | "Chemical antibodies"; synthetic, stable, and highly designable | The SELEX process for aptamer selection can be lengthy | Organophosphates, Carbamates [10] [3] |
| Enzymes (e.g., AChE) | Inhibition of catalytic activity | Excellent for class-selective detection of enzyme inhibitors | Low compound-specificity; detects all inhibitors of the enzyme | Organophosphates, Carbamates [10] [3] |
| Molecularly Imprinted Polymers (MIPs) | Size/shape-selective rebinding into synthetic cavities | High stability, reusable, and effective for small molecules | Can suffer from incomplete template removal and heterogeneous binding sites | Wide range, depending on the template [3] |
Experimental Protocols
The following protocols provide detailed methodologies for modifying SPCEs with different recognition elements and conducting pesticide detection assays.
Protocol 1: Fabrication of an Immunosensor for Organophosphate Detection
Principle: This protocol describes the development of a competitive electrochemical immunosensor using broad-spectrum antibodies and gold nanoparticle (AuNP) labels for the detection of organophosphate (OP) pesticides [71].
Materials:
- Screen-printed carbon electrodes (SPCEs)
- Broad-spectrum anti-OP antibodies
- Gold chloride (HAuCl₄), for AuNP synthesis
- Prussian blue (PB)
- Potassium ferrocyanide and ferricyanide
- Bovine Serum Albumin (BSA)
- Target OP pesticide standards (e.g., paraoxon, malathion)
- Electrochemical analyzer
Procedure:
- Synthesis of AuNP-Ab Conjugates: Synthesize AuNPs via the citrate reduction method. Mix the AuNP solution with the anti-OP antibodies and incubate to allow adsorption of the antibodies onto the AuNP surface via electrostatic interactions. Block remaining active sites on the AuNPs with BSA.
- Electrode Modification and Probe Co-deposition: Prepare a solution containing the AuNP-Ab conjugates, Prussian blue, and potassium ferricyanide/ferrocyanide. Deposit this mixture onto the working electrode of the SPCE using a one-step electrodeposition method (e.g., applying a constant potential for a set duration). This creates a nanocomposite film on the electrode surface.
- Competitive Immunoassay: Incubate the modified SPCE with a solution containing a mixture of the sample (or standard) and a fixed concentration of an enzyme-labeled OP pesticide (if using a direct competitive format) or simply with the sample for a label-free approach. In a competitive assay, the free OP pesticides in the sample and the labeled OP compete for the limited binding sites on the immobilized antibodies.
- Electrochemical Detection: After washing, perform electrochemical measurement. For the labeled format with Prussian blue, use Differential Pulse Voltammetry (DPV) to measure the reduction signal of Prussian blue. The signal is inversely proportional to the concentration of OP pesticide in the sample, as more OP binding leads to less enzyme-labeled OP binding and thus a lower catalytic current. For a label-free approach, Electrochemical Impedance Spectroscopy (EIS) can be used to measure the increase in charge-transfer resistance upon antibody-OP binding [71].
Protocol 2: Development of an Aptasensor for Carbamate Pesticides
Principle: This protocol outlines the construction of a label-free aptasensor where the binding of a carbamate pesticide to its specific DNA aptamer induces a conformational change, detectable via electrochemical impedance spectroscopy [10] [3].
Materials:
- SPCEs
- Carbamate-specific DNA aptamer sequence
- Methylene blue (redox probe)
- EDC/NHS (cross-linking agents)
- Carboxylated graphene oxide (carboxylated GO) or carboxylated multi-walled carbon nanotubes (MWCNTs)
- Target carbamate pesticide (e.g., carbofuran, carbaryl)
Procedure:
- SPCE Surface Activation: Drop-cast a dispersion of carboxylated GO or MWCNTs onto the working electrode of the SPCE and dry. This nanomaterial layer enhances the surface area and conductivity.
- Aptamer Immobilization: Activate the carboxyl groups on the nanomaterial surface using a mixture of EDC and NHS. Subsequently, incubate the electrode with the amine-modified DNA aptamer solution, allowing the formation of stable amide bonds, covalently immobilizing the aptamer.
- Blocking: Treat the electrode with a solution of BSA or ethanolamine to block any non-specific binding sites on the electrode surface.
- Target Incubation and Detection: Incubate the aptamer-modified SPCE with the sample or standard solution containing the target carbamate pesticide. Perform EIS measurements in a solution containing a [Fe(CN)₆]³⁻/⁴⁻ redox couple. The binding of the pesticide to the aptamer causes a conformational change and forms a complex on the surface, which increases the electron-transfer resistance (Rₑₜ). The change in Rₑₜ is proportional to the pesticide concentration [10].
Protocol 3: Enzymatic Sensor Based on Acetylcholinesterase Inhibition
Principle: This protocol details the use of acetylcholinesterase (AChE) inhibition for the class-selective detection of organophosphate and carbamate pesticides [10] [3].
Materials:
- SPCEs
- Acetylcholinesterase (AChE) enzyme
- Acetylthiocholine (ATCh) or acetylthiocholine iodide (ATCHI)
- Glutaraldehyde or Nafion (for enzyme immobilization)
- Phosphate buffer saline (PBS), pH 7.4
Procedure:
- Enzyme Immobilization: Mix AChE with a immobilization matrix such as a chitosan solution or a Nafion solution. Drop-cast this mixture onto the working electrode of the SPCE and allow it to dry. Alternatively, use cross-linking with glutaraldehyde to create a stable enzyme layer.
- Baseline Activity Measurement: Immerse the AChE-modified SPCE in a stirred PBS solution. Add a known concentration of ATCh substrate. Using Amperometry (AC) at a fixed applied potential (e.g., +0.5 V vs. Ag/AgCl), measure the steady-state current generated by the oxidation of the enzymatic product, thiocholine. This current represents the 100% enzyme activity (I₀).
- Enzyme Inhibition: Incubate the modified SPCE in a sample solution containing the pesticide for a fixed time (e.g., 10-15 minutes). The pesticide will inhibit the AChE enzyme.
- Inhibited Activity Measurement: Wash the electrode and repeat Step 2. Measure the new, lower steady-state current (Iᵢ).
- Quantification: The percentage of inhibition is calculated as % Inhibition = [(I₀ - Iᵢ) / I₀] × 100%. This value is proportional to the concentration of the inhibiting pesticide in the sample.
Table 2: Analytical Performance of Selected Recognition Element-Based Sensors
| Recognition Element | Target Pesticide | Electrochemical Technique | Limit of Detection (LOD) | Linear Range | Reference Context |
|---|---|---|---|---|---|
| Acetylcholinesterase | Organophosphates | Amperometry | ~0.38 pM (for class) | Not Specified | [3] |
| Antibodies | Organophosphates | DPV / EIS | Picomolar (pM) to Nanomolar (nM) range | Not Specified | [71] |
| Aptamer | Organophosphates | ECL / EIS | Picomolar (pM) level | Not Specified | [72] |
| Copper Oxide Nanozyme | Malathion | Colorimetric (Smartphone) | 0.08 mg/L | 0.1–5 mg/L | [3] |
| Molecularly Imprinted Polymer | Various | DPV | Nanomolar (nM) range | Not Specified | [3] |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Sensor Development
| Item / Reagent | Function / Application |
|---|---|
| Screen-Printed Carbon Electrodes (SPCEs) | Disposable, portable, and low-cost electrochemical transducer; the core platform for sensor development [10] [4]. |
| Gold Nanoparticles (AuNPs) | Nanomaterial for signal amplification; facilitates electron transfer and provides a surface for biomolecule immobilization [10] [71]. |
| Carbon Nanotubes (MWCNTs/SWCNTs) | Nanomaterial used to modify the electrode surface; increases electroactive surface area and enhances electrical conductivity [11] [10] [4]. |
| Graphene Oxide (GO) / Reduced GO | Nanomaterial with a high surface area and functional groups (e.g., -COOH) for covalent attachment of recognition elements [10] [4]. |
| Prussian Blue (PB) | An electrochemical redox mediator; used as a catalyst for H₂O₂ reduction and as a label in immunosensors [71]. |
| EDC/NHS Cross-linker | Activates carboxyl groups on nanomaterials or electrode surfaces for covalent immobilization of amine-containing biomolecules (antibodies, aptamers) [10]. |
| Acetylcholinesterase (AChE) | Enzyme used in inhibition-based biosensors for the detection of organophosphate and carbamate pesticides [10] [3]. |
| BSA (Bovine Serum Albumin) | A blocking agent used to passivate unused binding sites on the sensor surface, thereby reducing non-specific adsorption [10] [71]. |
Visual Workflows and Signaling Pathways
Immunosensor Development Workflow
Aptamer Binding and Signal Transduction
Enzymatic Inhibition Signaling Pathway
In the field of electrochemical biosensing for pesticide analysis, the biological recognition elements, such as natural enzymes, have long been a critical component. However, their inherent instability—sensitivity to environmental conditions, limited shelf life, and tendency to denature—poses significant challenges for reliable field deployment and long-term use [73] [3]. The emergence of nanozymes, a class of nanomaterials with intrinsic enzyme-like activities, presents a transformative solution to these limitations. Their unique properties, including exceptional structural stability, tunable catalytic activity, and cost-effectiveness, make them robust alternatives for integration into nanomaterial-modified screen-printed electrodes (SPEs) [74] [75]. This Application Note details the advantages of nanozymes and provides a standardized protocol for developing a nanozyme-based acetylcholinesterase (AChE)-mimic sensor for organophosphorus pesticide (OP) detection, a core methodology within the broader thesis research on advanced pesticide analysis platforms.
Comparative Advantages of Nanozymes
Nanozymes possess several distinct advantages over their natural counterparts, which are paramount for enhancing sensor stability and performance.
- Superior Physical and Chemical Stability: Unlike natural enzymes, which are proteins susceptible to denaturation under non-physiological conditions, nanozymes are inorganic nanomaterials. They maintain their catalytic activity under a wide range of pH levels and temperatures, and are resistant to proteolytic degradation [73] [76]. This ensures consistent performance in diverse and harsh environments where traditional biosensors would fail.
- Tunable and Multifunctional Catalysis: The catalytic activity of nanozymes can be finely tuned by engineering their size, shape, composition, and surface chemistry [73]. Furthermore, a single nanomaterial can exhibit multiple enzyme-like activities (e.g., peroxidase-like, oxidase-like, and catalase-like), enabling the design of sophisticated multi-enzyme cascade systems within a single sensing platform [77] [76].
- Ease of Production and Modification: Nanozymes can be synthesized in large quantities at a relatively low cost, bypassing the complex extraction and purification processes required for natural enzymes [73] [77]. Their surface is also readily modifiable with various functional groups or biomolecules (e.g., aptamers), which can enhance their specificity and catalytic efficiency [77].
Table 1: Quantitative Comparison of Natural Enzymes vs. Nanozymes
| Property | Natural Enzymes | Nanozymes |
|---|---|---|
| Catalytic Efficiency | High (e.g., (k{cat}/Km)) | Variable, can be optimized to rival natural enzymes [74] |
| Stability | Low (sensitive to pH, temperature, proteolysis) | High (stable under extreme conditions) [73] |
| Shelf Life | Weeks to months | Months to years [3] |
| Production Cost | High (complex purification) | Low (scalable synthesis) [73] [77] |
| Design Flexibility | Low (fixed structure/function) | High (tunable size, shape, composition) [73] |
| Multifunctionality | Typically single-activity | Often multi-enzymatic [77] |
Experimental Protocol: AChE-Mimic Nanozyme-Based Sensor for OP Detection
This protocol describes the development of an electrochemical sensor using a peroxidase (POD)-like nanozyme for the detection of organophosphorus pesticides (OPs) based on an inhibition mechanism.
Principle of Operation
Organophosphorus pesticides inhibit the activity of acetylcholinesterase (AChE). In this sensor, a POD-like nanozyme (e.g., CuO or Fe₃O₄ nanoparticles) replaces AChE in a mimicry system. The substrate acetylthiocholine (ATCh) is hydrolyzed by the nanozyme, producing thiocholine, which in the presence of H₂O₂ leads to an electrochemical signal. When OPs are present, they inhibit the nanozyme's catalytic activity, leading to a measurable decrease in the electrochemical signal, which is proportional to the pesticide concentration [3].
Materials and Reagents
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function/Description |
|---|---|
| Screen-Printed Electrode (SPE) | A disposable, portable, and mass-producible platform typically featuring a Carbon, Gold, or Platinum working electrode [1]. |
| CuO or Fe₃O₄ Nanoparticles | POD-like nanozyme that catalyzes the oxidation of substrates using H₂O₂ [3]. |
| Acetylthiocholine (ATCh) | Enzyme substrate. Hydrolyzed to produce thiocholine and acetic acid [3]. |
| Hydrogen Peroxide (H₂O₂) | Co-substrate for the POD-like reaction. |
| Nafion or Chitosan Solution | A polymer used to form a stable film on the SPE, entrapping the nanozymes and improving adhesion [1]. |
| Organophosphorus Pesticide Standard | Analytic (e.g., malathion, parathion). |
| Phosphate Buffered Saline (PBS) | Electrolyte solution for maintaining a stable pH during electrochemical measurement. |
Step-by-Step Procedure
Part A: Modification of the Screen-Printed Electrode
- Nanozyme Ink Preparation: Disperse 2 mg of synthesized CuO nanoparticles (NPs) in 1 mL of deionized water with 0.5% Nafion solution. Sonicate for 30 minutes to obtain a homogeneous ink.
- Electrode Modification: Pipette 5 µL of the nanozyme ink onto the clean working electrode surface of the SPE.
- Drying: Allow the modified electrode to dry at room temperature for 60 minutes, forming a stable nanozyme film.
- Storage: The modified SPEs (CuO/Nafion/SPE) can be stored dry at room temperature for several weeks prior to use.
Part B: Electrochemical Detection of Pesticides
- Baseline Measurement: Prepare a solution containing 0.1 M PBS (pH 7.4), 1.0 mM ATCh, and 2.0 mM H₂O₂. Immerse the modified SPE into the solution and record the amperometric current (I₀) at a fixed potential of +0.7 V (vs. Ag/AgCl reference on the SPE).
- Inhibition Step: Incubate the modified SPE in a sample solution containing the target OP pesticide for 15 minutes.
- Sample Measurement: After incubation, rinse the SPE gently with PBS. Re-immerse it in the ATCh/H₂O₂/PBS solution from Step 1 and record the amperometric current again (I).
- Data Analysis: The inhibition ratio is calculated as ((I₀ - I)/I₀ \times 100\%), which is then correlated to the pesticide concentration using a pre-established calibration curve.
Workflow and Signaling Pathway Visualization
The following diagram illustrates the experimental workflow and the signaling logic for the inhibition-based detection of pesticides.
Data Presentation and Analysis
The performance of the nanozyme-based sensor is characterized by its sensitivity, linear range, and limit of detection (LOD) for target pesticides. The following table summarizes typical performance metrics achievable with this protocol, based on data from recent literature.
Table 2: Analytical Performance of Representative Nanozyme-Based Sensors for Pesticide Detection
| Nanozyme Material | Detection Technique | Target Pesticide | Linear Range | Limit of Detection (LOD) | Stability / Shelf Life |
|---|---|---|---|---|---|
| Copper Oxide (CuO) NPs [3] | Colorimetric / Amperometric | Organophosphorus (e.g., Malathion) | 0.1 – 5 mg/L | 0.08 mg/L | > 4 weeks (room temperature) |
| Fe₃O₄ Nanoparticles [73] | Amperometric | Organophosphorus | – | – | High stability under extreme pH/temp [73] |
| DNA-templated Cu Nanoclusters [77] | Colorimetric | Various | – | – | Enhanced specificity and stability from DNA framework |
| Single-Atom Ce Nanozyme (SACe-N-C) [3] | Colorimetric | Organophosphorus | – | LOD in pM range | High turnover frequency and stability |
Troubleshooting and Notes
- Low Signal Response: Ensure the nanozyme ink is freshly prepared and well-dispersed. Aggregation of nanoparticles can significantly reduce catalytic activity. Check the activity of H₂O₂, as it decomposes over time.
- High Background Noise: Use high-purity reagents and deionized water. The SPE should be electrochemically cleaned in a blank buffer solution via cyclic voltammetry before modification if necessary.
- Poor Reproducibility: Standardize the drop-casting and drying process precisely. Consistent volume and homogeneous coverage of the nanozyme film are critical. Using an automated micropipette with a fixed angle for dispensing is recommended.
- Note on Specificity: While nanozymes are robust, their inherent specificity can be lower than that of natural enzymes. To enhance specificity, the sensor can be integrated with aptamers or molecularly imprinted polymers (MIPs) that selectively capture the target pesticide before it interacts with the nanozyme [3] [77].
The accurate and sensitive detection of pesticide residues is a critical requirement for ensuring food safety and environmental health. Within the broader context of developing nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, the optimization of key operational and fabrication parameters is paramount for achieving maximum sensor sensitivity and selectivity. Electrochemical methods are particularly promising for this application due to their affordability, simplicity, and suitability for field applications [11]. The performance of these electrochemical sensors is not solely dependent on the choice of nanomaterial but is profoundly influenced by a triad of interconnected parameters: the applied potential, the pH of the electrolyte medium, and the modification density of the nanostructured sensing layer. This application note provides a detailed, step-by-step protocol for systematically optimizing these parameters to enhance the sensitivity of SPE-based pesticide sensors.
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table catalogs the essential materials and reagents required for the fabrication and optimization of nanomaterial-modified SPEs for pesticide detection.
Table 1: Key Research Reagent Solutions and Materials
| Item | Function/Brief Explanation |
|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, portable platforms comprising working, reference, and counter electrodes; ideal for in-field analysis [11]. |
| Carbon Nanotubes (CNTs) | Nanomaterials used to modify the working electrode; enhance surface area, electrical conductivity, and electron transfer kinetics, leading to improved current response [11] [78]. |
| Gold Nanoparticles (AuNPs) | Nanomaterials acting as ion-electron transducers; offer excellent electrical conductivity, high stability, and simple production for enhanced signal stability [79]. |
| Calix[6]arene | A macrocyclic ionophore used in the sensing membrane; its large cavity allows for efficient binding with specific pesticide molecules, providing selectivity [79]. |
| Polyvinyl Chloride (PVC) | A polymer used as a structural matrix for the ion-selective membrane on the electrode surface. |
| 2-Nitrophenyl octyl ether (o-NPOE) | A plasticizer incorporated into the PVC membrane to provide flexibility and ensure proper ionophore mobility and function. |
| Potassium tetrakis(4-chlorophenyl)borate (K-TCPB) | An ionic additive in the sensing membrane that improves selectivity and reduces membrane resistance. |
| Phosphate Buffered Saline (PBS) | A common electrolyte solution used to maintain a stable pH during electrochemical measurements. |
| Pesticide Standard Solutions | Analytical standards of target analytes (e.g., MS222, p-nitrophenol, carbendazim) used for calibration and sensitivity assessment [11] [78]. |
Systematic optimization requires a structured approach to understanding how each parameter influences the sensor's output. The following table summarizes the key parameters and their optimized ranges as established in recent literature.
Table 2: Summary of Key Optimization Parameters for Pesticide Sensors
| Parameter | Impact on Sensor Performance | Optimal Range / Considerations | Supporting Evidence |
|---|---|---|---|
| Applied Potential | Directly influences the redox reaction of the target pesticide. An optimal potential maximizes Faradaic current while minimizing background noise. | Compound-specific; must be determined empirically via techniques like cyclic voltammetry (CV). For instance, a sensor for the pesticide fenobucarb showed high sensitivity using a specific potential on a modified SPCE [11]. | The choice dictates the driving force for electron transfer, critically affecting the signal-to-noise ratio [11]. |
| Solution pH | Affects the electrochemical activity of pesticide molecules and the surface charge of the nanomaterial modifier. | Varies by analyte; often slightly acidic to neutral (pH ~6-7.4). A pH of 7.4 was successfully used for the analysis of Mirabegron to mimic physiological conditions [79]. | pH can alter the protonation state of both the analyte and the sensor surface, impacting binding and electron transfer efficiency [79]. |
| Modification Density | Determines the number of active sites and the overall electroactive surface area. Too low limits sensitivity; too thick increases resistance and hinders electron transfer. | A monolayer or sub-monolayer coverage is often ideal. The performance of transducers like AuNPs and MWCNTs must be compared to identify the optimal loading for the best slope and potential stability [79]. | Higher loading can increase surface area and current response, but an excessively thick film can block electron transfer and reduce sensitivity [79] [78]. |
| Nanomaterial Type | Defines the fundamental electrocatalytic properties, conductivity, and surface area of the sensing interface. | Carbon nanotubes (CNTs), gold nanoparticles (AuNPs), metal oxides. Selection depends on the target pesticide. CNTs, for example, significantly increase surface area and current response [11] [78]. | The unique physicochemical properties of nanomaterials are key to enhancing sensor performance [11]. |
| Ionophore Type | Governs the molecular recognition and selectivity of the sensor towards a specific target. | Must exhibit high affinity for the target. Molecular docking simulations can predict affinity, e.g., Calix[6]arene showed a high docking score for Mirabegron [79]. | The host-guest chemistry between the ionophore and analyte is responsible for the desirable selectivity in sensors [79]. |
Detailed Experimental Protocols
Protocol 1: Electrode Modification with Nanomaterials
This protocol details the procedure for modifying a screen-printed carbon electrode with a nanomaterial layer, based on the drop-casting method [79].
Objective: To create a uniform, highly conductive, and catalytically active nanomaterial layer on the working electrode surface.
Materials:
- Screen-printed carbon electrodes (CH Instruments, 3 mm diameter)
- Multi-walled carbon nanotubes (MWCNT) or Gold Nanoparticle (GNP, 5 nm diameter) dispersion
- Xylene or Tetrahydrofuran (THF)
- Ultrasonic bath
Procedure:
- Dispersion Preparation: Weigh 10.0 mg of MWCNT powder and disperse in 1 mL of xylene. Alternatively, use a commercially available GNP dispersion.
- Sonication: Subject the dispersion to ultrasonic agitation for a minimum of 20 minutes to obtain a uniform, agglomerate-free suspension.
- Surface Cleaning: Clean the surface of the carbon SPE, if necessary, according to manufacturer instructions.
- Drop-Casting: Using a micropipette, deposit 10.0 µL of the nanomaterial dispersion directly onto the working electrode surface.
- Drying: Allow the modified electrode to dry overnight at room temperature, protected from dust.
- Quality Control: The resulting layer should appear uniform under visual inspection. Electrochemical characterization via Cyclic Voltammetry in a standard redox probe (e.g., 1 mM Potassium ferricyanide) can confirm successful modification by showing an increased peak current compared to an unmodified electrode.
Protocol 2: Systematic Optimization of pH and Applied Potential
This protocol outlines a coupled strategy for identifying the ideal pH and applied potential using cyclic voltammetry (CV) and differential pulse voltammetry (DPV).
Objective: To determine the pH and applied potential that yield the highest peak current and best-defined signal for the target pesticide.
Materials:
- Nanomaterial-modified SPE (from Protocol 1)
- Potentiostat/Galvanostat
- Phosphate buffer solutions (pH range 4.0 - 8.0)
- Standard solution of target pesticide (e.g., 100 µM)
Procedure:
- pH Profiling: a. Prepare a series of 0.1 M phosphate buffer solutions across a pH range (e.g., 4.0, 5.0, 6.0, 7.0, 8.0). b. Add a fixed concentration of the pesticide standard to each buffer solution. c. Using the modified SPE, perform CV scans (e.g., from -0.5 V to +0.8 V) in each solution. d. Record the peak potential (Ep) and peak current (Ip) for the pesticide's oxidation/reduction. e. Plot Ip and Ep versus pH. The pH that produces the maximum Ip is typically selected for subsequent experiments. The relationship between Ep and pH can reveal the number of protons involved in the electrochemical reaction.
- Applied Potential Optimization via DPV: a. Using the optimal pH identified in Step 1, prepare a solution containing the pesticide standard. b. Employ DPV, as it offers better sensitivity and resolution for analytical applications compared to CV [78]. c. Acquire DPV curves, ensuring the potential window encompasses the oxidation/reduction peak identified in the CV scans. d. The characteristic peak in the DPV voltammogram, often called an "electrochemical fingerprint," identifies the optimal applied potential for that specific pesticide on your modified sensor [78]. This potential should be used for all subsequent quantitative calibration experiments.
Workflow Visualization: Sensor Fabrication and Optimization Pathway
The following diagram illustrates the logical workflow integrating the protocols for sensor fabrication, optimization, and deployment.
Analytical Validation and Performance Benchmarking Against Standard Methods
The accurate quantification of pesticide residues is paramount for ensuring food safety and environmental health. Within the context of a broader thesis on nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, this document details the critical analytical performance metrics—detection limits, linear ranges, and sensitivity—reported in recent, seminal studies. SPEs provide a robust, disposable, and cost-effective electroanalytical platform, ideal for in-field and point-of-care testing [4]. Their performance is significantly enhanced through strategic modification with nanomaterials, which improve electrocatalytic activity, increase surface area, and facilitate electron transfer, thereby achieving the sensitivity required for detecting trace-level pesticide residues in complex matrices [80] [81] [32]. This note consolidates quantitative data into structured tables and provides the detailed experimental protocols necessary for the replication and advancement of these sensor platforms by researchers and scientists in drug development and analytical chemistry.
Performance Metrics of Recent Sensor Platforms
The following table summarizes the key analytical performance metrics for a selection of highly effective nanomaterial-modified SPEs developed for pesticide detection. These platforms exemplify the synergy between advanced nanomaterials and electrochemical transduction.
Table 1: Analytical Performance of Nanomaterial-Modified SPEs for Pesticide Detection
| Target Pesticide | Sensor Platform | Detection Technique | Linear Range | Limit of Detection (LOD) | Sensitivity | Application in Real Samples |
|---|---|---|---|---|---|---|
| Carbaryl (CAR) [80] | MXene/CNF/SPE | Square Wave Voltammetry (SWV) | 2.0 × 10⁻⁶ to 3.9 × 10⁻⁵ mol L⁻¹ | 5.2 × 10⁻⁷ mol L⁻¹ | Not Specified | Environmental samples |
| Glyphosate [82] | Cu²⁺/4-MBA/AuNPs/SPE | Square Wave Voltammetry (SWV) | 5 to 100 nM | 1.65 nM | Not Specified | Tap water |
| Chlorpyrifos [83] | CdS/SPCE | Differential Pulse Voltammetry (DPV) | 5–80 nM; 100–1000 nM | 0.0106 nM | 22.95 × 10⁻⁴ mA/nM·cm² (5-80 nM) | Water and soil |
| Acetamiprid (AAP) [84] | AgNPs/SPE | Electrochemical SERS (EC-SERS) | 0.1 to 1000 μM | 0.04 μM | Not Specified | Brassica chinensis L. |
Detailed Experimental Protocols
This protocol outlines the development of a versatile sensor for simultaneous detection.
- 1. Electrode Modification: Disposable screen-printed electrodes are modified by dispensing a homogeneous suspension containing MXene (Ti₃C₂Tx) and carbon nanofibers (CNF) directly onto the surface of the carbon working electrode. The modified electrode is then dried under ambient conditions or in an oven at a mild temperature (e.g., 50 °C) to form a stable film.
- 2. Electrochemical Measurement: All electrochemical measurements are performed using a standard three-electrode system: the modified SPE (working electrode), its integrated reference (e.g., Ag/AgCl), and counter electrodes. A potentiostat/galvanostat system controlled by appropriate software (e.g., GPES) is used.
- 3. Analysis Procedure:
- Supporting Electrolyte: Use a 0.10 mol L⁻¹ phosphate buffer solution (PBS) at pH 7.0 as the supporting electrolyte.
- Electrochemical Characterization: Characterize the electrode using Cyclic Voltammetry (CV) in a potential range of 0 to 1.2 V at a scan rate of 50 mV s⁻¹.
- Analyte Quantification: Employ Square Wave Voltammetry (SWV) under optimized parameters (e.g., frequency, amplitude, step potential). The oxidation peaks for carbaryl are monitored.
- Calibration: Construct an analytical curve by spiking standard solutions of carbaryl into the electrolyte and recording the SWV response. The peak current is plotted against concentration to establish a linear range of 2.0 × 10⁻⁶ to 3.9 × 10⁻⁵ mol L⁻¹.
- 4. Real Sample Analysis: For environmental sample analysis (e.g., water), employ a standard addition method or dilute the sample with the phosphate buffer solution. Recovery rates close to 100% validate the method's accuracy.
This protocol describes a highly specific biosensor leveraging a self-assembled monolayer on gold nanoparticles.
- 1. Electrode Modification:
- AuNPs Electrodeposition: Gold nanoparticles (AuNPs) are electrodeposited onto the bare carbon working electrode of an SPE by applying a constant potential or using cyclic voltammetry in a solution of HAuCl₄.
- SAM Formation: Immerse the AuNPs/SPE in an ethanol solution containing 4-mercaptobenzoic acid (4-MBA) to form a self-assembled monolayer (SAM) via Au-S bonds.
- Cu²⁺ Immobilization: Incubate the 4-MBA/AuNPs/SPE in a copper ions (Cu²⁺) solution. The Cu²⁺ immobilizes onto the monolayer via coordination bonding, finalizing the Cu²⁺/4-MBA/AuNPs/SPE biosensor.
- 2. Biosensor Characterization: Confirm successful modification at each step using Scanning Electron Microscopy (SEM), Energy-Dispersive X-ray Spectroscopy (EDX), and electrochemical techniques like Electrochemical Impedance Spectroscopy (EIS).
- 3. Analysis Procedure:
- Detection Principle: The detection is based on the specific interaction between glyphosate and the immobilized Cu²⁺, which causes a measurable change in the electrochemical signal.
- Square Wave Voltammetry: Use SWV to measure the peak current. The presence of glyphosate causes an inhibition of the peak current, and the inhibition ratio is proportional to the glyphosate concentration.
- Quantification: A linear relationship is established between the glyphosate concentration and the peak current inhibition ratio over a range of 5-100 nM, achieving an LOD of 1.65 nM.
- 4. Real Sample Analysis: Spike tap water samples with known glyphosate concentrations. The biosensor demonstrates excellent recovery rates (89.84%–107.48%), confirming its practicality.
This protocol combines the specificity of SERS with the controlled enhancement of electrochemistry.
- 1. Substrate Preparation: Synthesize silver nanoparticles (AgNPs) (~40 nm) via hydroxylamine reduction. Modify the SPE by simply drop-casting the AgNPs colloidal solution onto the working electrode surface to create the AgNPs/SPE substrate.
- 2. EC-SERS Measurement:
- Setup: Place the AgNPs/SPE in a standard three-electrode electrochemical cell containing the analyte solution (acetamiprid).
- Potential Application: Apply an optimal constant potential of -0.7 V (vs. the integrated Ag/AgCl reference) to the working electrode. This potential enhances the adsorption of AAP molecules onto the AgNP surface, significantly boosting the SERS signal.
- SERS Detection: While the potential is applied, acquire the Raman spectrum of the analyte. The intensity of characteristic Raman peaks is used for quantification.
- 3. Analysis Procedure:
- Calibration: A wide linear range from 0.1 μM to 1000 μM is achieved, with an LOD of 0.04 μM, which is 7-fold lower than conventional SERS without the applied potential.
- Validation: The results from real vegetable samples (Brassica chinensis L.) show a strong correlation (R² = 0.9760) with those obtained from HPLC-MS, demonstrating high accuracy.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagents and Materials for Sensor Fabrication and Analysis
| Reagent/Material | Function in Research | Example Use Case |
|---|---|---|
| Screen-Printed Electrodes (SPEs) | Disposable, portable platform integrating working, reference, and counter electrodes; facilitates mass production and miniaturization [4]. | Base transducer for all sensor designs. |
| MXene (Ti₃C₂Tx) | 2D nanomaterial providing high electrical conductivity, hydrophilicity, and a large specific surface area to enhance electrocatalytic activity [80]. | Used with CNF to modify SPE for carbaryl detection [80]. |
| Carbon Nanofibers (CNFs) | Cylindrical nanostructures that provide edge plane defects, promoting electron transfer and increasing the electroactive surface area [80]. | Composite material in MXene/CNF/SPE [80]. |
| Gold Nanoparticles (AuNPs) | Provide high electrocatalytic activity, biocompatibility, and a surface for forming self-assembled monolayers (SAMs) via thiol-gold chemistry [82] [85]. | Electrodeposited on SPE for glyphosate aptasensor [82]. |
| Silver Nanoparticles (AgNPs) | Used for high SERS enhancement due to strong surface plasmon resonance; serve as an excellent substrate for EC-SERS [84]. | Drop-cast on SPE for acetamiprid detection [84]. |
| Cadmium Sulfide (CdS) Nanostructures | Semiconductor nanomaterial with good electrocatalytic properties and a high surface area for biomolecule immobilization and electron transfer [83]. | Hydrothermally synthesized to modify SPCE for chlorpyrifos sensing [83]. |
| Aptamers | Single-stranded DNA/RNA oligonucleotides serving as synthetic biorecognition elements; offer high selectivity, stability, and regenerability compared to antibodies [85]. | Recognition element for targets like glyphosate and carbendazim [82] [85]. |
| Phosphate Buffer Solution (PBS) | A common supporting electrolyte in electrochemistry that maintains a stable pH, crucial for reproducible redox reactions [80]. | Used as the electrolyte in the MXene/CNF sensor (pH 7.0) [80]. |
Workflow Diagrams for Sensor Operation
The following diagrams illustrate the core operational principles and experimental workflows for two primary types of sensors discussed.
The analysis of pesticide residues represents a critical challenge in environmental monitoring, food safety, and public health. Traditional chromatographic methods like high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) and gas chromatography-mass spectrometry (GC-MS) have established themselves as reference techniques for pesticide detection due to their high sensitivity, selectivity, and reliability [86] [87]. These methods enable precise quantification of pesticide residues across various matrices, complying with stringent international regulations such as the European Union's Maximum Residue Levels (MRLs) [87].
In recent years, screen-printed electrode (SPE) technology has emerged as a powerful complementary approach, particularly when modified with nanomaterials and biological recognition elements [1] [10]. These sensors offer remarkable advantages including portability, rapid analysis, cost-effectiveness, and potential for field-deployable applications [10] [2]. However, to gain acceptance in analytical science, these novel platforms require rigorous validation against established standard methods. This protocol details comprehensive procedures for correlating data from nanomaterial-modified SPEs with traditional chromatographic techniques, ensuring the reliability and credibility of electrochemical sensing strategies for pesticide analysis.
Establishing the Validation Framework
Key Validation Parameters
Method validation ensures that an analytical procedure is suitable for its intended purpose. The correlation study between nanomaterial-modified SPEs and standard chromatographic methods should be designed to evaluate the following parameters:
- Accuracy: Assessed through recovery studies by spiking known pesticide concentrations into real samples and comparing measured values between techniques.
- Precision: Evaluated as both intra-day (repeatability) and inter-day (intermediate precision) variations.
- Sensitivity: Determined by comparing limits of detection (LOD) and quantification (LOQ) across methods.
- Linearity and Range: Established by analyzing calibration curves over the method's dynamic range.
- Selectivity: Verified by testing against potentially interfering compounds present in sample matrices.
Regulatory Considerations
Validation protocols should align with established guidelines such as the FDA Reviewer Guidance: Validation of Chromatographic Methods and the SANTE/11312/2021 v2 document [88] [87]. These frameworks provide standardized approaches for method validation, ensuring regulatory acceptance and interoperability of data across different laboratories and platforms.
Table 1: Key Validation Parameters and Target Performance Criteria
| Parameter | Evaluation Method | Target Criteria | Reference Method |
|---|---|---|---|
| Accuracy | Recovery studies using spiked samples | 70-120% recovery with RSD <15% | HPLC-MS/MS [87] |
| Precision | Repeated measurements (n≥5) | Intra-day & inter-day RSD <15% | HPLC-MS/MS [87] |
| LOD | Signal-to-noise ratio (S/N=3) | Method-dependent; e.g., 0.005 mg/kg for LC-MS/MS | Documented reference values [87] |
| LOQ | Signal-to-noise ratio (S/N=10) | Method-dependent; meets regulatory needs | Documented reference values [87] |
| Linearity | Coefficient of determination (R²) | R² > 0.990 | HPLC-MS/MS calibration [87] |
| Selectivity | Interference studies | <±20% signal deviation | Chromatographic separation [86] |
Experimental Protocols
Protocol 1: HPLC-MS/MS Reference Method for Pesticide Analysis
This protocol adapts validated methods from recent literature for determining pesticide residues in complex matrices [86] [87].
Materials and Reagents:
- HPLC-grade solvents: Acetonitrile, methanol, water
- Analytical standards: Target pesticides (e.g., organophosphates, carbamates, isothiazolinones)
- Additives: Formic acid, ammonium acetate or formate
- Sample preparation: QuEChERS extraction kits or alternative materials
Procedure:
Sample Preparation:
- Homogenize representative sample (1.0 ± 0.1 g).
- Extract using appropriate solvent (e.g., acetonitrile with 1% formic acid).
- Employ cleanup step using d-SPE (e.g., C18, PSA, magnesium sulfate).
- Concentrate extract under gentle nitrogen stream and reconstitute in initial mobile phase.
HPLC-MS/MS Analysis:
- Chromatography:
- Column: C18 reverse-phase (100 × 2.1 mm, 1.8-2.7 μm)
- Mobile phase: (A) Water with 0.1% formic acid; (B) Acetonitrile with 0.1% formic acid
- Gradient: 5-95% B over 10-15 minutes
- Flow rate: 0.3-0.5 mL/min
- Injection volume: 1-10 μL
- Mass Spectrometry:
- Ionization: Electrospray ionization (ESI) in positive/negative mode
- Operation: Multiple reaction monitoring (MRM)
- Optimize source parameters for each pesticide (capillary voltage, cone voltage, collision energy)
- Chromatography:
Quantification:
- Prepare matrix-matched calibration standards (0.010-0.500 mg/L)
- Use internal standards where applicable for improved accuracy
Figure 1: HPLC-MS/MS Reference Method Workflow
Protocol 2: Nanomaterial-Modified SPE Sensor for Pesticide Detection
This protocol details the development and application of nanomaterial-modified SPEs for electrochemical pesticide detection, with emphasis on correlation with reference methods [1] [10].
Materials and Reagents:
- Screen-printed electrodes: Carbon, gold, or platinum working electrodes
- Nanomaterials: Graphene oxide, carbon nanotubes, metal nanoparticles
- Biorecognition elements: Acetylcholinesterase (AChE), antibodies, aptamers
- Electrochemical probes: e.g., [Fe(CN)₆]³⁻/⁴⁻
- Buffer solutions: Phosphate buffer (0.1 M, pH 7.4), acetate buffer
Sensor Preparation Procedure:
Electrode Modification:
- Clean SPE surface electrochemically or chemically.
- Deposit nanomaterial suspension (e.g., 5-10 μL of graphene oxide or CNT dispersion) on working electrode.
- Dry under ambient temperature or infrared light.
- Immobilize biorecognition element (enzyme, antibody, or aptamer) via cross-linking or physical adsorption.
Electrochemical Measurements:
- Technique: Cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), or differential pulse voltammetry (DPV)
- Parameters:
- For CV: Scan rate 50-100 mV/s, potential range -0.2 to +0.8 V
- For EIS: Frequency range 0.1-100,000 Hz, amplitude 10 mV
- For DPV: Pulse amplitude 50 mV, pulse width 50 ms
- Measurement: Record signal before and after pesticide exposure
Detection Mechanisms:
- Enzymatic inhibition: Measure decreased enzyme activity in presence of pesticides
- Direct electrochemistry: Detect electroactive pesticides
- Affinity biosensing: Monitor binding events through impedance changes
Figure 2: Nanomaterial-Modified SPE Sensor Preparation and Measurement Workflow
Correlation Study Design and Data Analysis
Sample Set Design
For comprehensive method correlation, analyze a diverse set of samples:
- Fortified samples: Spiked with known pesticide concentrations across linear range
- Real-world samples: Agricultural products, environmental waters, food extracts
- Blank samples: Verify absence of target analytes and matrix effects
- Proficiency testing materials: If available for quality assurance
Statistical Analysis for Method Correlation
Regression Analysis:
- Perform linear regression (SPE results vs. HPLC-MS/MS results)
- Calculate slope, intercept, and coefficient of determination (R²)
- Ideal correlation: Slope = 1, intercept = 0, R² > 0.95
Bland-Altman Analysis:
- Plot difference between methods against their mean
- Establish limits of agreement (mean difference ± 1.96 SD)
- Identify potential biases between methods
Statistical Tests:
- Paired t-test for significant differences between methods
- F-test for comparing variances between methods
Table 2: Comparative Analytical Performance of SPE vs. Chromatographic Methods
| Analyte Class | Detection Technique | Linear Range | LOD | Recovery (%) | Analysis Time | Reference |
|---|---|---|---|---|---|---|
| Organophosphates | AChE-Inhibition SPE | 0.1-100 μg/L | 0.05 μg/L | 85-115 | <30 min | [10] |
| Organophosphates | HPLC-MS/MS | 0.01-50 μg/L | 0.005 μg/L | 87-114 | >60 min | [87] |
| Carbamates | Enzymatic SPE | 1-500 μg/L | 0.5 μg/L | 82-118 | <30 min | [10] |
| Carbamates | GC-MS/MS | 0.05-100 μg/L | 0.02 μg/L | 88-116 | >60 min | [10] |
| Isothiazolinones | Immunosensor SPE | 0.5-200 μg/L | 0.2 μg/L | 80-120 | <20 min | [1] |
| Isothiazolinones | HPLC-MS/MS | 0.01-0.5 mg/L | 0.003 mg/L | 87-115 | >60 min | [86] |
Case Study: Isothiazolinone Detection in Consumer Products
A recent study demonstrates the correlation approach for isothiazolinone detection [86]. Researchers developed an HPLC-ESI-MS/MS method for determining isothiazolinone migration from children's sports protectors into artificial sweat, achieving:
- Linearity: 0.010-0.500 mg/L with R² > 0.9990
- Quantification limits: 0.7-3.0 μg
- Recovery: 87.2-114.8% with RSDs <10%
- Precision: Intra-day and inter-day RSDs <8%
This validated chromatographic method serves as an excellent reference for correlating with SPE-based approaches for the same analytes. Similar validation criteria should be applied to SPE sensors targeting these compounds.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for Method Validation Studies
| Reagent/Material | Function/Application | Specification Requirements | Example Suppliers |
|---|---|---|---|
| HPLC-MS/MS Grade Solvents | Mobile phase preparation | Low UV absorbance, high purity, LC-MS compatible | Fisher Scientific, Sigma-Aldrich |
| Certified Reference Standards | Calibration & quantification | Certified purity, traceability to reference materials | NIST, ERA, LGC Standards |
| Nanomaterials for SPE Modification | Electrode surface enhancement | Defined particle size, functionalized surfaces | Sigma-Aldrich, NanoComposix |
| Screen-Printed Electrodes | Electrochemical sensing platform | High reproducibility, customizable designs | Metrohm, DropSens, Zimmer & Peacock |
| Enzymes (AChE, BChE, Tyr) | Biosensor recognition elements | High specific activity, stability | Sigma-Aldrich, Roche |
| QuEChERS Kits | Sample preparation & cleanup | Matrix-specific formulations | Agilent, Thermo Scientific |
| Artificial Sweat/Saliva | Migration studies | Standardized composition | Pickering Laboratories, in-house preparation |
Troubleshooting and Technical Notes
Common Challenges and Solutions
- Matrix Effects: Different sample matrices can affect SPE and HPLC-MS/MS performance differently. Use matrix-matched calibration standards and standard addition methods to compensate.
- Sensor Fouling: Complex samples can degrade SPE performance. Implement regeneration protocols between measurements and use protective membranes.
- Cross-reactivity: SPE biosensors may show interference from related compounds. Evaluate selectivity with structurally similar compounds.
- Reproducibility: SPE batch-to-batch variations can occur. Characterize multiple electrodes from different production batches.
Data Interpretation Guidelines
- Acceptable Correlation: R² > 0.95 with slope between 0.8-1.2 indicates good method agreement.
- Systematic Bias: Consistent over- or under-estimation by SPE may require calibration adjustment.
- Outliers: Investigate analytical errors or sample-specific interferences for discordant results.
- Dynamic Range: Ensure compared methods have overlapping linear ranges for meaningful correlation.
This protocol provides a comprehensive framework for validating nanomaterial-modified screen-printed electrodes against established chromatographic reference methods for pesticide analysis. The systematic correlation approach ensures that innovative electrochemical sensors meet the rigorous demands of analytical science while leveraging their inherent advantages of portability, rapid analysis, and cost-effectiveness [1] [10]. As SPE technology continues to advance, these validation protocols will be essential for bridging novel sensing platforms with regulatory acceptance and practical implementation in environmental monitoring, food safety, and public health protection.
Within the broader research on nanomaterial-modified screen-printed electrodes (SPEs) for pesticide analysis, the transition from controlled laboratory buffers to complex real-world samples represents a critical validation step. The matrix effects from fruits, vegetables, biological fluids, and environmental samples present significant challenges, including fouling agents, interfering compounds, and variable pH/ionic strength, which can severely impact sensor accuracy and reliability [1] [10]. This application note provides a detailed protocol for evaluating the performance of nanomaterial-modified SPEs in these complex matrices, with structured quantitative data and experimental methodologies to guide researchers and scientists in drug development and environmental monitoring.
The following tables summarize the analytical performance of nanomaterial-modified SPEs across various sample types and pesticide classes, highlighting their versatility in real-sample applications.
Table 1: Performance of Nanomaterial-Modified SPEs in Fruit and Vegetable Matrices
| Pesticide Class | Specific Analytes | Sample Matrix | Sensor Configuration | LOD | Recovery (%) | RSD (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Organophosphorus (OPs) | Parathion-methyl, Fenitrothion | Apple, Spinach, Tomato | AChE/ChOx-MWCNT/SPE | 0.05-0.1 nM | 85.2-112.8 | 3.2-8.5 | [89] [10] |
| Carbamates (CPs) | Carbaryl, Carbofuran | Cabbage, Orange | AChE/ZnONPs/SPE | 0.1-0.5 nM | 88.5-105.3 | 4.1-9.7 | [10] [3] |
| Organochlorines (OCs) | α-BHC, γ-BHC | Carrot, Potato | Immunosensor/AuNPs/SPE | 0.01-0.05 ppb | 90.1-108.7 | 5.2-12.1 | [89] [2] |
| Pyrethroids (PYs) | Permethrin, Deltamethrin | Strawberry, Grape | MIP/rGO/SPE | 0.1-0.3 ppb | 92.4-106.9 | 4.8-11.3 | [89] [3] |
| Herbicides (Triazines) | Atrazine, Simazine | Wheat, Corn | Aptasensor/MoS2/SPE | 0.05-0.2 ppb | 94.2-103.5 | 3.8-7.9 | [90] [3] |
Table 2: Performance in Biological and Environmental Samples
| Sample Type | Pesticide Class | Key Analytes | Sensor Configuration | LOD | Recovery (%) | Linear Range | Ref. |
|---|---|---|---|---|---|---|---|
| Human Serum/Urine | Organophosphorus | Chlorpyrifos, Malathion | AChE/Prussian Blue/SPE | 0.1-0.5 pM | 91-109 | 1 pM - 1 µM | [10] [91] |
| Lake/River Water | Organophosphorus, Carbamates | Parathion, Carbofuran | OPH Enzyme/CNF/SPE | 0.05-0.2 ppb | 85-112 | 0.1-100 ppb | [10] [91] |
| Agricultural Soil | Neonicotinoids | Imidacloprid, Thiamethoxam | Antibody/Fe3O4/SPE | 0.01-0.03 ppb | 89-107 | 0.05-50 ppb | [90] [3] |
| Groundwater | Herbicides | Atrazine, Diuron | MIP/CQDs/SPE | 0.02-0.08 ppb | 93-104 | 0.1-80 ppb | [3] [2] |
Detailed Experimental Protocols
Protocol 1: Sample Preparation for Fruits and Vegetables
This protocol adapts the multiplug filtration clean-up (m-PFC) method for use with SPE-based sensors, effectively removing pigments and organic acids that interfere with electrochemical detection [89].
Reagents and Materials:
- Acetonitrile (chromatographic grade)
- Multi-walled carbon nanotubes (MWCNTs) m-PFC column (e.g., B-5 or C-5 type, 750-800 mg/6 mL)
- Buffer extraction solution
- Ultrapure water
- Ceramic homogenizer
- Nitrogen evaporator
Procedure:
- Homogenization: Weigh 15.0 ± 0.1 g of homogenized fruit/vegetable sample into a 50 mL centrifuge tube.
- Extraction: Add 15 mL of acetonitrile and 6 g of buffer extraction salts. Shake vigorously for 1 minute using a high-speed vortex mixer.
- Centrifugation: Centrifuge at 4200 rpm for 5 minutes at 4°C to separate phases.
- Purification: Transfer 8 mL of the supernatant to an m-PFC column. Allow it to pass through via gravity flow (~2-3 minutes).
- Concentration: Collect the eluate and evaporate to near dryness under a gentle nitrogen stream at 40°C.
- Reconstitution: Reconstitute the residue in 1 mL of appropriate electrolyte (e.g., 0.1 M PBS, pH 7.4) for electrochemical analysis.
- Analysis: The purified extract is now ready for measurement with the modified SPE.
Critical Notes:
- The m-PFC method utilizes MWCNTs to effectively adsorb interfering pigments and sugars, achieving recovery rates of 83.8-112.8% for 37 pesticides across 8 different matrices [89].
- This method significantly reduces matrix effects compared to traditional solid-phase extraction (SPE), with detection limits ranging from 0.0001 to 0.03 μg kg−1 for most pesticides [89].
Protocol 2: Electrochemical Measurement Using Modified SPEs
This protocol details the electrochemical detection of pesticides using acetylcholinesterase (AChE)-inhibition based biosensors, suitable for organophosphorus and carbamate pesticides [10] [3].
Apparatus:
- Potentiostat/Galvanostat
- Screen-printed electrode (Carbon, Gold, or Platinum working electrode)
- Nanomaterial-modified SPE (e.g., MWCNT/Chitosan, AuNPs/rGO)
- Micro-pipettes
Immobilization Procedure (AChE-based Biosensor):
- Surface Modification: Deposit 5 μL of nanomaterial suspension (e.g., 1 mg/mL MWCNTs in DMF) on the working electrode and dry at 40°C.
- Enzyme Immobilization: Apply 5 μL of AChE solution (0.5 U/μL in 0.1 M PBS, pH 7.4) and incubate for 30 minutes at 25°C.
- Stabilization: Rinse gently with PBS to remove unbound enzyme.
- Storage: Store the modified SPE at 4°C when not in use.
Measurement Procedure (Inhibition Mode):
- Baseline Measurement: Place the modified SPE in 0.1 M PBS (pH 7.4) containing 1.0 mM acetylthiocholine (ATCh). Record the amperometric current (I₀) at +0.65 V vs. Ag/AgCl for 60 seconds.
- Inhibition Step: Incubate the SPE in the sample extract containing pesticide for 10 minutes at 25°C.
- Post-Inhibition Measurement: Re-immerse the SPE in fresh ATCh/PBS solution and record the current (I).
- Quantification: Calculate inhibition percentage as: % Inhibition = [(I₀ - I)/I₀] × 100
Calibration:
- Prepare calibration curves by plotting % inhibition vs. logarithm of pesticide concentration.
- The inhibition mechanism relies on phosphorylation of the serine residue in the AChE active center, blocking acetylcholine hydrolysis [10] [3].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for SPE-Based Pesticide Detection
| Reagent/Material | Function/Description | Application Examples |
|---|---|---|
| MWCNT m-PFC Columns | Multi-walled carbon nanotubes for purification; effectively removes pigments, sugars, organic acids | Sample clean-up for fruits/vegetables prior to SPE analysis [89] |
| Acetylcholinesterase (AChE) | Enzyme inhibition-based recognition element for OPs and carbamates | Biosensor fabrication for neurotoxic pesticides [10] [3] |
| Organophosphorus Hydrolase (OPH) | Enzyme for catalytic detection of OPs; hydrolyzes pesticides directly | Direct measurement of OPs in environmental samples [10] [91] |
| Gold Nanoparticles (AuNPs) | Enhances electron transfer, provides immobilization platform | Signal amplification in immunosensors and aptasensors [90] [2] |
| Reduced Graphene Oxide (rGO) | High surface area, excellent electrical conductivity | Electrode modification for enhanced sensitivity [10] [92] |
| Molecularly Imprinted Polymers (MIPs) | Synthetic receptors with high stability and selectivity | Biomimetic sensors for herbicides and pyrethroids [3] [2] |
| Prussian Blue | Electron mediator for hydrogen peroxide detection | Enzyme-based biosensors with low working potential [10] [91] |
| Phosphate Buffered Saline (PBS) | Electrolyte solution for electrochemical measurements | Standard medium for electrochemical measurements [10] [2] |
Experimental Workflow and Signaling Pathways
Figure 1: Overall experimental workflow for pesticide detection in real samples using modified SPEs, covering sample preparation to data validation.
Figure 2: Signaling pathway for enzyme inhibition-based detection of organophosphorus and carbamate pesticides, showing the mechanism of signal reduction upon pesticide binding.
The protocols and data presented herein demonstrate that nanomaterial-modified SPEs provide reliable performance across diverse real sample matrices, achieving detection limits sufficient for monitoring compliance with regulatory standards such as the EU's maximum residue limits (MRLs) of 0.1 µg/L for individual pesticides [10]. The integration of advanced nanomaterials with appropriate sample preparation techniques enables researchers to overcome matrix effects and obtain accurate pesticide quantification in complex samples.
The simultaneous detection of multiple pesticide residues, or multiplexing, addresses a critical analytical challenge in food safety and environmental monitoring. Conventional methods often struggle with the reality that food samples frequently contain complex mixtures of chemical contaminants rather than isolated single compounds [93]. Multiplex detection strategies provide significant advantages for high-throughput screening by reducing analysis time, lowering costs per sample, and offering a more comprehensive safety profile of tested products [93]. The foundation of these multiplexing approaches lies in two primary strategies: using broadly specific recognition elements that can interact with multiple related analytes, or utilizing inherent characteristics of pesticides that can be measured without target-specific receptors [93].
Within the specific context of nanomaterial-modified screen-printed electrodes (SPEs), multiplexing capability is significantly enhanced through strategic electrode design and modification. SPEs provide an ideal platform for multiplexed biosensing due to their low cost, portability, and ease of modification [1] [25]. The incorporation of nanomaterials such as noble metal nanoparticles, carbon nanotubes, and graphene derivatives dramatically improves electrochemical performance by increasing surface area, enhancing electron transfer kinetics, and providing abundant sites for immobilizing biological recognition elements [25] [32]. This synergistic combination of SPEs with tailored nanomaterials enables the development of compact, disposable devices capable of rapidly quantifying multiple pesticide residues in complex matrices with sensitivity rivaling traditional laboratory techniques [49] [25].
Recognition Strategies for Multiplexed Detection
Broadly Specific Recognition Elements
The development of recognition elements with cross-reactivity toward multiple targets is a cornerstone of multiplexed pesticide detection. These elements provide the necessary binding affinity for several structurally related analytes within a single assay format.
Generic Antibodies: These antibodies are generated by immunizing hosts with "general-structure" haptens that preserve the common steric and electronic features of an entire pesticide class [93]. For instance, generic haptens containing an O,O-diethyl thiophosphate moiety have been used to produce antibodies broadly specific to numerous organophosphorus pesticides (OPs) [93]. Computer-assisted molecular modeling at the three-dimensional level has significantly improved the rational design of these haptens, enabling more predictable cross-reactivity patterns without extensive trial-and-error experimentation [93].
Aptamers: Single-stranded DNA or RNA molecules obtained through Systematic Evolution of Ligands by EXponential enrichment (SELEX) offer several advantages for multiplexing, including thermal stability, ease of modification, and the ability to be selected for multiple targets [93] [3]. Their synthetic nature and compatibility with various nanomaterials make them particularly suitable for integration with SPE platforms.
Molecularly Imprinted Polymers (MIPs): These synthetic biomimetic receptors contain tailor-made binding cavities complementary to the shape and functional groups of target molecules [93] [3]. MIPs demonstrate exceptional stability under harsh chemical and thermal conditions where biological receptors would denature, offering significant advantages for field-deployable sensors intended for use in diverse environments [93].
Enzyme Systems: Enzymes such as acetylcholinesterase (AChE) and tyrosinase naturally respond to multiple pesticides within the same chemical class through inhibition mechanisms [25] [94]. For example, AChE inhibition forms the basis for detecting organophosphates and carbamates collectively, providing a group-specific detection approach rather than compound-specific quantification [25].
Spatial Multiplexing with Array-based Approaches
An alternative to broadly specific receptors involves creating sensor arrays with multiple discrete detection zones, each functionalized with different specific recognition elements. This spatial multiplexing approach enables true multi-analyte detection in a single device. Research demonstrates the feasibility of this strategy, such as the development of a multiplex immunochromatographic electrochemical biosensor (IEB) capable of simultaneous detection of three organophosphate insecticides (chlorpyrifos, parathion, and fenitrothion) and three herbicides (atrazine, cyanazine, and hydroxytriazine) [49]. In such configurations, each test zone contains a different capture element, allowing for parallel quantification while maintaining high specificity. The miniaturized format of SPEs makes them particularly amenable to such array designs, where multiple working electrodes can be patterned on a single chip substrate, each modified with different receptors and potentially even different nanomaterials optimized for specific detection chemistries [1] [25].
Nanomaterial-enhanced Screen-Printed Electrode Platforms
Key Nanomaterials and Their Functions
The analytical performance of SPE-based sensors for pesticide detection is profoundly enhanced through nanomaterial modifications. These materials contribute unique physicochemical properties that significantly improve detection sensitivity, selectivity, and stability.
Table 1: Key Nanomaterials for SPE Modification in Pesticide Detection
| Nanomaterial Category | Specific Examples | Key Functions and Properties | Representative Applications |
|---|---|---|---|
| Noble Metal Nanoparticles | Gold nanoparticles (AuNPs), Silver nanoparticles (AgNPs), Platinum nanoparticles (PtNPs) | Enhanced electrical conductivity, catalytic activity, large surface area, excellent biocompatibility for biomolecule immobilization [25] [32] | AuNPs with AChE for organophosphorus detection [32]; Pt-based bimetal nanoparticles as peroxidase mimics [49] |
| Carbon-based Nanomaterials | Multi-walled carbon nanotubes (MWCNTs), Graphene, Reduced graphene oxide (rGO) | High electrical conductivity, large specific surface area, π-π stacking interactions with aromatic compounds, promotion of electron transfer reactions [25] [94] | MWCNT-chitosan-AuNP composites for OP detection [94]; Graphene-modified SPEs [25] |
| Nanohybrid Materials | Pt-Au, Pt-Pd, Pt-Co bimetal nanoparticles, Carbon nanotube-metal nanoparticle composites | Synergistic effects combining properties of individual components, enhanced catalytic activity, improved stability, tailored electronic properties [49] [32] | Pt-based bimetal nanoparticles for multiplex immunosensing [49]; MWCNT-AuNP composites [94] |
| Quantum Dots | CdTe QDs, Carbon quantum dots | Unique optical and electronic properties, size-tunable fluorescence, photoinduced electron transfer capabilities | CdTe QD aerogels in fluorescent microfluidic sensors for OPs [3] |
Transduction Mechanisms in Electrochemical Detection
The integration of nanomaterials with SPEs enhances various electrochemical transduction mechanisms that form the basis of pesticide detection:
Voltammetric Techniques: Methods including cyclic voltammetry (CV), differential pulse voltammetry (DPV), and square wave voltammetry (SWV) measure current resulting from oxidation or reduction reactions of electroactive species. Nanomaterials increase electrode surface area and enhance electron transfer kinetics, resulting in significantly improved sensitivity and lower detection limits [25]. DPV and SWV are particularly valuable for their ability to minimize charging currents, thereby enhancing the faradaic current signal relative to background.
Amperometric Detection: This technique measures current at a fixed potential over time and is widely used in enzyme-based biosensors. The excellent electrocatalytic properties of nanomaterials like platinum nanoparticles and carbon nanotubes enable sensitive detection of enzymatic products at lower applied potentials, reducing interference from other electroactive species in complex samples [25] [94].
Electrochemical Impedance Spectroscopy (EIS): EIS measures changes in charge transfer resistance at the electrode-electrolyte interface, providing a label-free detection method particularly suitable for affinity-based sensors (aptasensors, immunosensors). Nanomaterials significantly enhance the surface area available for biorecognition events, amplifying the impedance change upon target binding [25].
Experimental Protocols and Methodologies
Protocol 1: Fabrication of Nanomaterial-modified SPEs
Objective: Prepare reproducible, high-performance screen-printed electrodes modified with nanomaterial composites for multiplexed pesticide detection.
Materials:
- Commercial carbon SPEs (e.g., DRP-110 from Metrohm DropSens)
- Gold nanoparticles (AuNPs, 10-20 nm diameter)
- Multi-walled carbon nanotubes (MWCNTs)
- Chitosan (medium molecular weight)
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
- Glutaraldehyde (2.5% solution in water)
Procedure:
- MWCNT Suspension Preparation: Disperse 1.0 mg of MWCNTs in 1.0 mL of 0.5% chitosan solution (prepared in 1% acetic acid). Sonicate the mixture for 30 minutes using a probe sonicator to achieve a homogeneous black suspension.
- Electrode Pretreatment: Electrochemically clean the carbon SPE by performing 10 cycles of cyclic voltammetry from 0 to +1.0 V in 0.5 M H₂SO₄ at a scan rate of 100 mV/s.
- Nanomaterial Modification: Deposit 5 μL of the MWCNT-chitosan suspension onto the working electrode area and allow to dry at room temperature for 1 hour.
- AuNP Modification: Drop-cast 3 μL of AuNP colloidal solution onto the MWCNT-modified electrode and dry under ambient conditions.
- Cross-linking: Apply 2 μL of 2.5% glutaraldehyde solution to the modified surface and allow to react for 30 minutes to create a stable cross-linked matrix.
- Washing: Rinse the modified SPE thoroughly with PBS (pH 7.4) to remove unbound reagents.
- Storage: Store the modified SPEs at 4°C in a desiccator until use.
Validation: Characterize the modified electrode surface using scanning electron microscopy to confirm uniform nanomaterial distribution. Electrochemically validate using cyclic voltammetry in 5 mM Fe(CN)₆³⁻/⁴⁻ to observe enhanced peak currents and reduced peak separation compared to unmodified SPEs [94].
Protocol 2: Enzyme-based Multiplexed Detection of Organophosphates and Carbamates
Objective: Simultaneously detect multiple organophosphorus and carbamate pesticides via acetylcholinesterase inhibition using nanomaterial-modified SPEs.
Materials:
- Nanomaterial-modified SPEs (from Protocol 1)
- Acetylcholinesterase (AChE, Type VI-S, from electric eel)
- Acetylthiocholine iodide (ATCh) or acetylthiocholine chloride (ATCl)
- Potassium ferricyanide (K₃[Fe(CN)₆])
- PBS (0.1 M, pH 7.4)
- Standard pesticide solutions (parathion, chlorpyrifos, carbaryl)
Procedure:
- Enzyme Immobilization: Deposit 3 μL of AChE solution (0.5 U/μL in PBS, pH 7.4) onto the nanomaterial-modified working electrode. Incubate for 1 hour at 4°C in a humidified chamber to prevent evaporation.
- Blocking: Apply 5 μL of 1% bovine serum albumin (BSA) solution for 30 minutes to block nonspecific binding sites. Rinse gently with PBS.
- Baseline Measurement: Perform amperometric measurement in electrochemical cell containing 2.0 mM ATCh and 1.0 mM K₃[Fe(CN)₆] in PBS. Apply a constant potential of +0.45 V vs Ag/AgCl and record the steady-state current (I₀).
- Inhibition Step: Incubate the biosensor for 10 minutes in sample solution containing target pesticides. Rinse gently with PBS to remove unbound inhibitors.
- Post-inhibition Measurement: Measure current (Iᵢ) again in fresh substrate solution using the same amperometric parameters.
- Quantification: Calculate percentage inhibition using the formula: % Inhibition = [(I₀ - Iᵢ)/I₀] × 100.
- Calibration: Construct calibration curves by plotting % inhibition versus logarithm of pesticide concentration for each target analyte.
Performance Metrics: This protocol typically achieves detection limits of 0.01-0.1 μg/L for organophosphates like paraoxon-ethyl, with two linear dynamic ranges (0.01-10 μg/L and 10-100 μg/L) [94]. The biosensor retains approximately 83% of initial enzyme activity after 49 days of dry storage at 4°C.
Protocol 3: Immunosensor-based Multiplexed Detection Using Spatial Arrays
Objective: Simultaneously detect multiple specific pesticides using an antibody-based array on a single SPE platform.
Materials:
- SPEs with multiple working electrodes (e.g., 4-electrode configuration)
- Pesticide-specific antibodies (anti-chlorpyrifos, anti-atrazine, anti-parathion)
- Protein A or G solution
- BSA
- Pt-based bimetal nanoparticle labels (Pt-Au, Pt-Pd)
- Hydrogen peroxide (H₂O₂)
- TMB (3,3',5,5'-tetramethylbenzidine) substrate
Procedure:
- Electrode Patterning: Design SPE with multiple working electrodes; each will be functionalized with a different capture antigen.
- Surface Activation: Activate each working electrode surface by electrochemical pretreatment or oxygen plasma treatment to introduce functional groups.
- Capture Immobilization: Spot 1 μL of different pesticide-protein conjugate solutions (10 μg/mL in PBS) onto specific working electrodes. Allow to adsorb for 2 hours at room temperature in a humidified chamber.
- Blocking: Treat the entire electrode array with 1% BSA for 1 hour to block nonspecific binding sites.
- Competitive Assay: Incubate the sensor with 50 μL of sample solution mixed with antibody-bimetal nanoparticle conjugates for 15 minutes.
- Washing: Rinse thoroughly with PBS containing 0.05% Tween 20 to remove unbound conjugates.
- Electrochemical Detection: Add H₂O₂ substrate and measure the electrocatalytic current at each working electrode using amperometry at -0.2 V vs Ag/AgCl.
- Data Analysis: Quantify each pesticide based on the inverse relationship between signal intensity and analyte concentration in the competitive assay format.
Performance Metrics: This approach demonstrated simultaneous detection of six pesticides (three organophosphates and three herbicides) with limits of detection significantly below EPA tolerances. The use of Pt-based bimetal nanoparticles provided excellent peroxidase-like catalysis and signal amplification [49].
Research Reagent Solutions
Table 2: Essential Research Reagents for Multiplexed Pesticide Detection
| Reagent Category | Specific Examples | Function in Experimental Workflow | Key Characteristics |
|---|---|---|---|
| Recognition Elements | Generic antibodies, Aptamers, Molecularly imprinted polymers, Acetylcholinesterase | Molecular recognition of target pesticides; provide assay specificity and cross-reactivity patterns | Antibodies offer high affinity; aptamers have thermal stability; MIPs have excellent chemical stability; enzymes provide class-specific detection [93] [3] |
| Nanomaterials | AuNPs, AgNPs, MWCNTs, Graphene, Pt-based bimetallic nanoparticles | Signal amplification, enhanced electron transfer, increased surface area for bioreceptor immobilization | Noble metals offer conductivity and catalytic properties; carbon materials provide large surface area; hybrids create synergistic effects [49] [32] |
| Electrode Systems | Carbon SPEs, Gold SPEs, Ceramic-based SPEs, Multi-electrode arrays | Sensor platform providing the electrochemical transduction interface | Disposable, low-cost, portable, amenable to mass production and various surface modifications [1] [25] |
| Electrochemical Substrates/Mediators | Ferricyanide, H₂O₂/TMB, Acetylthiocholine, Catechol | Generate measurable electrochemical signals through redox reactions | Ferricyanide is a common diffusional mediator; H₂O₂/TMB is used with peroxidase-like nanozymes; acetylthiocholine is enzyme substrate [25] [94] |
| Immobilization Matrices | Chitosan, Nafion, Glutaraldehyde, BSA | Stabilize and attach recognition elements to electrode surfaces | Chitosan offers biocompatibility and film-forming ability; glutaraldehyde provides cross-linking; BSA blocks nonspecific sites [94] |
Analytical Performance and Validation
The performance of multiplexed detection platforms using nanomaterial-modified SPEs has been rigorously validated against established reference methods. For example, an AChE-based biosensor incorporating MWCNT-AuNP nanocomposites demonstrated excellent correlation with HPLC for paraoxon-ethyl detection in spinach samples, confirming method reliability [94]. Similarly, a multiplex immunosensing device successfully detected six pesticides in fruits, vegetables, and groundwater with sensitivity exceeding regulatory requirements [49].
Critical performance metrics include:
- Detection Limits: Typically range from ng/L to low μg/L, significantly below maximum residue limits (MRLs) established by regulatory agencies [32] [94]
- Linear Dynamic Range: Often spans 2-3 orders of magnitude, with some sensors exhibiting dual linear ranges (e.g., 0.01-10 μg/L and 10-100 μg/L) [94]
- Reproducibility: Relative standard deviations (RSD) generally below 10% for inter-electrode and intra-assay precision [25]
- Stability: Nanomaterial-modified biosensors typically retain >80% initial activity after one month of proper storage [94]
- Analysis Time: Rapid detection ranging from 10-30 minutes, significantly faster than chromatographic methods requiring extensive sample preparation [49] [3]
Table 3: Performance Comparison of Multiplexed Detection Approaches
| Detection Platform | Target Pesticides | Linear Range | Detection Limit | Assay Time | Real Sample Application |
|---|---|---|---|---|---|
| Enzyme Inhibition (AChE) | Organophosphates, Carbamates (class detection) | 0.01-100 μg/L | 0.01-0.1 μg/L | 15-20 min | Spinach, fruits [94] |
| Multiplex Immunosensor | Chlorpyrifos, Parathion, Fenitrothion, Atrazine, Cyanazine, Hydroxytriazine | Varies by analyte | Below EPA tolerances | ~15 min | Fruits, vegetables, groundwater [49] |
| Aptamer-based Sensor | Chlorpyrifos | - | 36 ng/L | - | Apple, pak choi [32] |
| Nanozyme-based Sensor | Organophosphates | 0.1-5 mg/L | 0.08 mg/L | ~10 min | Fruits, vegetables [3] |
Multiplexed detection of pesticide residues using nanomaterial-modified screen-printed electrodes represents a significant advancement in analytical science for food safety and environmental monitoring. The integration of broadly specific recognition elements with the enhanced electrochemical properties afforded by nanomaterials creates powerful analytical tools that combine the sensitivity of laboratory methods with the practicality of field-deployment. As research continues to refine these technologies, focusing on improved receptor design, advanced nanomaterial synthesis, and miniaturized instrumentation, these multiplexed platforms are poised to become increasingly important for comprehensive pesticide screening programs, ultimately contributing to enhanced food safety and environmental protection.
The integration of nanomaterial-modified screen-printed electrodes (SPEs) represents a transformative advancement in electrochemical biosensing for pesticide analysis [2] [95]. These sensors leverage the unique properties of nanomaterials to achieve remarkable sensitivity, portability, and cost-effectiveness, positioning them as ideal candidates for on-site environmental and food safety monitoring [4] [95]. However, the path from laboratory innovation to commercially viable and widely trusted technology is contingent upon solving critical challenges in reproducibility and reliability [2] [96].
A significant barrier to commercialization is the inconsistency in nanomaterial synthesis and electrode modification processes, which can lead to variable sensor performance across different production batches and laboratories [4] [96]. Furthermore, carbon-based nanomaterials are known to interfere with standard assay methods, potentially yielding false results if not properly controlled [97]. This application note details standardized protocols and analytical frameworks designed to systematically evaluate and enhance the reproducibility and reliability of these promising sensing platforms, providing a pathway toward robust commercial application.
The table below summarizes performance data from recent studies on nanomaterial-modified SPEs for pesticide detection, highlighting key metrics relevant to reproducibility and reliability assessment.
Table 1: Performance Metrics of Selected Nanomaterial-Modified SPEs for Pesticide Detection
| Target Pesticide | Nanomaterial Used | Sensor Type | Limit of Detection (LOD) | Linear Range | Reported Stability | Reference |
|---|---|---|---|---|---|---|
| Chlorpyrifos | Gold Nanoparticles (AuNPs) | Immunosensor | 0.01 ng/mL | 0.5–20 ng/mL | 95% signal after 4 weeks | [95] |
| Organophosphates | 3D Graphene/CuO Nanoflowers | Enzymatic (AChE) Biosensor | 0.35 pM | 1 pM – 0.1 nM | Not specified | [95] |
| Methamidophos | Genetically Modified AChE | Amperometric Biosensor | Attomolar range | Not specified | Not specified | [95] |
| Malathion | Polydopamine-Gold NPs, Exonuclease I | Aptasensor | 0.033 pM | 0.1 pM – 1 nM | Good reproducibility (RSD < 5%) | [95] |
| Various Pesticides | Carbon Nanotubes (CNTs), Graphene (GR) | Electrochemical (Bio)sensors | Varies by design | Varies by design | Highly dependent on modification stability | [95] |
Experimental Protocols
Protocol for Fabrication of Nanomaterial-Modified SPEs
Title: Standardized Fabrication of Nanomaterial-Modified Screen-Printed Electrodes. Objective: To establish a reproducible method for modifying carbon SPEs with graphene oxide (GO) and gold nanoparticles (AuNPs) to create a stable sensing platform [4] [95].
Materials:
- Commercial carbon SPEs (e.g., Metrohm)
- Graphene oxide (GO) dispersion (1 mg/mL in deionized water)
- Hydrogen tetrachloroaurate(III) trihydrate (HAuCl₄·3H₂O)
- Sodium citrate dihydrate
- Phosphate Buffered Saline (PBS), 0.1 M, pH 7.4
- Ethanol (absolute)
- Ultrasonic bath
- Centrifuge
- Electrochemical workstation
Procedure:
- SPE Pre-treatment:
- Cycle the bare carbon SPE in 0.1 M H₂SO₄ via cyclic voltammetry (CV) from -0.2 V to +0.6 V (vs. Ag/AgCl) for 10 cycles at a scan rate of 100 mV/s to activate the surface [4].
- Rinse the electrode thoroughly with deionized water and dry under a gentle nitrogen stream.
Graphene Oxide Modification:
- Dilute the GO dispersion to 0.5 mg/mL in a 1:1 ethanol/water solution.
- Dispense 5 µL of the diluted GO suspension onto the clean working electrode surface.
- Allow the electrode to dry overnight at room temperature in a desiccator. This results in a GO-modified SPE (GO/SPE).
Gold Nanoparticle (AuNP) Synthesis & Deposition:
- Prepare AuNPs by the Turkevich method: Heat 100 mL of 1 mM HAuCl₄ to boiling under vigorous stirring. Rapidly add 2.5 mL of 38.8 mM sodium citrate solution. Continue heating and stirring until the solution turns deep red (≈15 minutes). Cool to room temperature [98].
- Characterize the AuNP solution by UV-Vis spectroscopy; a peak at ≈520 nm confirms formation.
- Drop-cast 5 µL of the synthesized AuNP solution onto the GO/SPE surface.
- Dry for 2 hours at room temperature. The final electrode is designated AuNP/GO/SPE.
Quality Control Check:
- Characterize the modified electrode using CV and Electrochemical Impedance Spectroscopy (EIS) in a 5 mM [Fe(CN)₆]³⁻/⁴⁻ solution.
- A successful modification is indicated by a decreased charge transfer resistance (Rₑₜ) in EIS and enhanced peak currents in CV compared to the bare SPE [4].
Protocol for Inter-laboratory Reproducibility Study
Title: Inter-laboratory Study for Reproducibility Assessment of Modified SPEs. Objective: To quantify the inter-laboratory and intra-batch reproducibility of the fabrication and analytical performance of nanomaterial-modified SPEs [96].
Materials:
- Identical batches of raw materials (SPEs, GO, HAuCl₄, etc.) distributed to all participating laboratories.
- Standardized pesticide solutions (e.g., Chlorpyrifos, 100 µg/mL stock in methanol).
- Ferricyanide/ferrocyanide redox probe solution.
- Multi-laboratory cohort (minimum of 3 independent labs).
Procedure:
- Fabrication Phase:
- Each participating laboratory fabricates a set of n=15 AuNP/GO/SPEs using the standardized protocol in Section 3.1.
- Laboratories record batch numbers, environmental conditions (temperature, humidity), and any deviations.
Electrochemical Characterization:
- Each lab performs EIS on all 15 electrodes in the ferricyanide/ferrocyanide solution.
- Record the charge transfer resistance (Rₑₜ) for each electrode.
- Calculate the intra-laboratory relative standard deviation (RSD) for the Rₑₜ values.
Analytical Performance Assessment:
- Using a common calibration protocol, each lab measures the amperometric response of n=5 electrodes to a standard Chlorpyrifos solution (e.g., 10 ng/mL).
- Record the average response and standard deviation for each lab's set of electrodes.
Data Analysis and Reporting:
- The coordinating center collates Rₑₜ values and analytical responses from all labs.
- Statistical analysis is performed to calculate:
- Inter-laboratory RSD: The variation of the average Rₑₜ (or analytical response) across different labs.
- Total RSD: The overall variation combining within-lab and between-lab variability.
- A total RSD of ≤15% for the key performance metric (e.g., Rₑₜ or analytical signal) is typically targeted for acceptable reproducibility in analytical chemistry [96].
The Scientist's Toolkit: Essential Research Reagents and Materials
The successful development and reliable reproduction of nanomaterial-modified SPEs depend on a core set of materials and reagents. The following table details these essential components and their critical functions in the sensor fabrication and testing workflow.
Table 2: Key Research Reagent Solutions and Materials for SPE-Based Pesticide Sensors
| Item | Function/Description | Critical Parameters for Reliability |
|---|---|---|
| Carbon Screen-Printed Electrodes (SPEs) | Disposable three-electrode cell (Working, Reference, Counter); platform for modification [2] [4]. | Substrate material (e.g., PVC, ceramic), ink composition (graphite, CNTs), and geometric consistency. |
| Graphene Oxide (GO) | A 2D carbon nanomaterial that provides a high-surface-area scaffold for anchoring nanoparticles and biomolecules, enhancing electron transfer [4] [95]. | Degree of oxidation, number of layers, sheet size, and dispersion concentration. |
| Gold Nanoparticles (AuNPs) | Plasmonic nanoparticles that significantly enhance electrochemical signals and serve as a platform for immobilizing biorecognition elements (e.g., antibodies, aptamers) [98] [95]. | Particle size distribution, morphology (spherical, rods), surface charge (zeta potential), and concentration. |
| Acetylcholinesterase (AChE) Enzyme | A common biorecognition element in enzymatic biosensors for organophosphate and carbamate pesticides, which inhibit its activity [99] [95]. | Enzyme source (electric eel, genetically modified), specific activity, and storage stability. |
| Aptamers | Single-stranded DNA or RNA oligonucleotides that bind to specific pesticide targets with high affinity; used in aptasensors [95]. | Nucleotide sequence, purity, and folding conditions. |
| Ferricyanide/Ferrocyanide Redox Probe | A standard electrochemical probe ([Fe(CN)₆]³⁻/⁴⁻) used to characterize the electrode surface before and after modification via EIS and CV [4]. | Consistent molarity and purity to ensure reliable baseline measurements. |
| Standard Pesticide Solutions | Certified reference materials used for sensor calibration and validation [95]. | Purity, concentration, and solvent matrix. Must be traceable to international standards. |
Reliability and Interference Testing Protocol
Title: Assessing Reliability and Mitigating Nanomaterial-Based Interference. Objective: To evaluate the operational stability of the modified SPEs and identify/correct for potential false signals caused by nanomaterial interference [97].
Materials:
- Fabricated AuNP/GO/SPEs from Protocol 3.1.
- Target pesticide solutions.
- Control solutions (blank, potentially interfering compounds).
- LIVE/DEAD assay kit (or other relevant biological assay if testing cytotoxicity).
Procedure:
- Stability Testing:
- Perform CV or amperometric measurements on a single electrode over 50 consecutive cycles/scans.
- Calculate the RSD of the signal to assess short-term operational stability.
- Store a batch of electrodes at 4°C and measure their response to a standard analyte weekly for one month. A signal retention >90% indicates good long-term stability.
- Interference Testing:
- Control Experiments: Always run parallel control experiments with the modified SPEs that lack the specific biorecognition element (e.g., no enzyme, no aptamer).
- Standard Addition Method: Spike known concentrations of the analyte into a real sample matrix and measure the recovery. Recovery rates between 80-120% are generally acceptable.
- For Cytotoxicity Assays (if applicable): As demonstrated with carbon nanomaterials, implement specific gating strategies in flow cytometry and include additional controls (e.g., nanomaterials alone, dyes with nanomaterials) to account for quenching or autofluorescence [97].
Achieving commercial viability for nanomaterial-modified SPEs in pesticide monitoring is a multifaceted challenge that extends beyond high sensitivity. It demands a rigorous, standardized framework for assessing and ensuring reproducibility and reliability across laboratories and production batches. The protocols and data analysis frameworks presented here—covering standardized fabrication, inter-laboratory studies, interference checks, and stability testing—provide a foundational approach to tackling these challenges. By adopting such systematic quality control measures, researchers and developers can significantly enhance the credibility of their sensor platforms, accelerating their transition from promising lab prototypes to reliable tools for ensuring food safety and environmental health.
The economic analysis of analytical techniques is a critical consideration for modern laboratories, particularly in fields requiring routine monitoring such as pesticide analysis. Traditional laboratory methods, including chromatography-based techniques,, , have long been considered the gold standard for accuracy and sensitivity. However, their operational costs, maintenance requirements, and need for specialized personnel present significant economic challenges. The emergence of nanomaterial-modified screen-printed electrodes (SPEs) offers a compelling alternative that balances analytical performance with substantial economic benefits. This application note provides a detailed cost-benefit analysis and experimental protocols for implementing SPE-based sensors within a research context focused on pesticide analysis, demonstrating their economic advantages without compromising analytical rigor.
Quantitative Economic Analysis: SPEs vs. Conventional Techniques
Market Growth and Industry Validation
The significant market growth for SPE-based technologies provides strong indirect evidence of their economic viability. The global screen-printed electrodes market, valued at approximately USD 652.46 million in 2025, is projected to cross USD 1.5 billion by 2035, expanding at a compound annual growth rate (CAGR) of 8.7% during the forecast period [100]. This robust growth trajectory, particularly in the medical diagnostics sector which accounts for approximately 40% of the SPE market, signals strong industry confidence in the cost-effectiveness of this technology [101]. The carbon-based SPE segment is poised to account for more than 58.2% of the market share by 2035, largely due to increasing requirements for point-of-care testing and decentralized diagnostic solutions [100].
Direct Cost Comparison
Table 1: Comprehensive Cost-Benefit Analysis: SPEs vs. Conventional Techniques
| Parameter | Nanomaterial-Modified SPEs | Conventional Laboratory Techniques (HPLC/GC-MS) |
|---|---|---|
| Initial Instrumentation Cost | $5,000 - $15,000 [41] | $50,000 - $150,000 [81] |
| Cost Per Analysis | $1 - $5 (disposable electrodes) [100] | $50 - $200 (solvents, standards, columns) [81] |
| Sample Preparation Time | Minutes (minimal preparation) [1] | Hours (extraction, cleanup, derivation) [35] |
| Analysis Time | Seconds to minutes [41] | 10-30 minutes per sample [81] |
| Personnel Requirements | Minimal training required [41] | Highly trained technicians [81] |
| Portability & Field Use | Excellent [102] | Limited to laboratory settings [81] |
| Manufacturing Scalability | High-throughput production possible [100] | Limited to batch production [81] |
| Detection Limit | nM-μM range (suitable for regulatory compliance) [1] | pM-nM range (superior sensitivity) [81] |
The economic advantage of SPEs becomes particularly evident when considering the total cost of ownership. While conventional techniques like HPLC and GC-MS offer superior sensitivity with detection limits in the pM-nM range [81], SPE technology provides sufficient sensitivity (nM-μM range) for many regulatory compliance scenarios at a fraction of the cost [1]. The disposability of SPEs eliminates cleaning procedures and prevents cross-contamination, reducing analysis time and labor costs [41]. Furthermore, the minimal solvent requirements of electrochemical detection compared to chromatographic methods significantly reduce both costs and environmental impact [35].
Experimental Protocols for SPE-Based Pesticide Analysis
Fabrication of Nanomaterial-Modified Screen-Printed Electrodes
Principle: Screen-printing technology deposits successive layers of conductive and insulating inks onto various substrates (typically ceramic or plastic), creating a three-electrode system (working electrode, reference electrode, and counter electrode) integrated on a single chip [41]. Modification with nanomaterials enhances sensitivity, selectivity, and stability for pesticide detection [1].
Materials:
- Screen-printing apparatus (printer, screens, squeegee)
- Carbon-based conductive ink (graphite, graphene, carbon nanotubes)
- Insulating polymer ink
- Silver/silver chloride ink for reference electrode
- Plastic or ceramic substrates
- Nanomaterials for modification (gold nanoparticles, carbon nanotubes, graphene oxide, metal oxides)
Procedure:
- Substrate Preparation: Clean the substrate surface (typically PVC, polycarbonate, or ceramic) to ensure proper ink adhesion.
- Conductive Layer Deposition: Using a patterned screen, deposit conductive carbon ink to form the electrode pathways and contacts. Cure according to manufacturer specifications (typically 60-120°C for 10-30 minutes).
- Reference Electrode Formation: Print Ag/AgCl ink onto the designated area to form the pseudo-reference electrode. Cure at appropriate temperature.
- Insulating Layer Application: Print insulating dielectric polymer ink to define the exact electrode area and create contact insulation. Cure to complete the basic SPE structure.
- Nanomaterial Modification: a. Drop-Casting Method: Prepare nanomaterial dispersion (e.g., 1 mg/mL graphene oxide in water). Apply precise volume (typically 5-10 μL) to the working electrode surface. Allow to dry at room temperature or mild heating [1]. b. Electrodeposition: For metal nanoparticles, immerse SPE in solution containing metal ions (e.g., 1 mM HAuCl₄) and apply controlled potential to deposit nanoparticles directly onto the electrode surface [67].
- Quality Control: Perform electrochemical characterization using cyclic voltammetry in standard redox probes (e.g., 1 mM ferrocenemethanol or potassium ferricyanide) to verify performance [103].
Troubleshooting Tips:
- If reproducibility issues occur, ensure consistent ink viscosity and printing parameters.
- For irregular nanomaterial deposition, use surfactant-free dispersions and optimize solvent evaporation conditions.
- If reference electrode instability is observed, ensure proper curing and consider using commercial SPEs as a reliable baseline [41].
Electrochemical Detection of Organophosphorus Pesticides
Principle: This protocol utilizes acetylcholinesterase (AChE) enzyme inhibition for selective detection of organophosphorus pesticides (OPs). Pesticides inhibit AChE activity, reducing enzymatic hydrolysis of acetylthiocholine substrate, which is electrochemically measured [1].
Materials:
- Nanomaterial-modified SPEs (e.g., CNT-modified or AuNP-modified)
- Acetylcholinesterase enzyme (AChE)
- Acetylthiocholine chloride substrate
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
- Standard pesticide solutions (parathion, malathion, chlorpyrifos)
- Electrochemical analyzer (potentiostat)
Procedure:
- Enzyme Immobilization: Incubate SPE with 10 μL AChE solution (0.5 U/mL) for 30 minutes at 4°C. Rinse gently with PBS to remove unbound enzyme.
- Baseline Measurement: Record amperometric response in PBS containing 0.5 mM acetylthiocholine chloride at applied potential of +0.5 V vs. Ag/AgCl.
- Enzyme Inhibition: Incubate AChE-modified SPE with pesticide sample for 10-15 minutes. Rinse with PBS.
- Post-Inhibition Measurement: Record amperometric response under identical conditions as step 2.
- Quantification: Calculate percentage inhibition = [(I₀ - I₁)/I₀] × 100%, where I₀ and I₁ are currents before and after inhibition. Compare with calibration curve from standard solutions.
Analysis Conditions:
- Technique: Amperometry or Square Wave Voltammetry
- Potential: +0.5 V vs. Ag/AgCl (for thiocholine oxidation)
- Buffer: 0.1 M PBS, pH 7.4
- Temperature: 25°C
- Incubation time: 10-15 minutes
Validation:
- Test with known pesticide standards to establish calibration curves.
- Evaluate cross-reactivity with other pesticide classes.
- Assess recovery in spiked real samples (fruits, vegetables) [35].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for SPE-Based Pesticide Sensors
| Material/Reagent | Function | Application Notes |
|---|---|---|
| Carbon Nanotubes (CNTs) | Enhance electron transfer, increase surface area | Disperse in DMF or water (0.5-1 mg/mL) for electrode modification [1] |
| Gold Nanoparticles (AuNPs) | Electrocatalytic properties, biocompatible surface for biomolecule immobilization | Synthesize by citrate reduction; electrodeposit on SPE [67] |
| Graphene Oxide (GO) | High surface area, rich functional groups for modification | Reduce electrochemically after deposition for enhanced conductivity [81] |
| Acetylcholinesterase (AChE) | Biological recognition element for organophosphorus pesticides | Immobilize via cross-linking with glutaraldehyde or physical adsorption [1] |
| Cellulose Nanocrystals (CNCs) | Biocompatible substrate for nanoparticle stabilization | Provides stable matrix for metal nanoparticle dispersion on electrode surface [67] |
| Chinese Shellac Biopolymer | Sustainable binder for carbon-based conductive inks | Eco-friendly alternative to synthetic polymers; compatible with carbon black [103] |
Application Workflow and Signaling Pathways
The following diagram illustrates the complete experimental workflow for SPE-based pesticide detection, from electrode fabrication through to analytical measurement:
The economic advantages of nanomaterial-modified SPEs for pesticide analysis are substantial and multifaceted. When considering total analytical costs including instrumentation, consumables, personnel, and time efficiency, SPE-based methodologies can reduce operational expenses by 60-80% compared to conventional laboratory techniques [100]. The disposability of SPEs addresses contamination concerns while the minimal sample preparation requirements significantly increase analytical throughput. For research and monitoring applications where ultra-trace (ppb) detection is not essential, SPE technology represents an optimal balance of performance, practicality, and economic efficiency. Future developments in nanomaterial integration, manufacturing automation, and multiplexing capabilities will further enhance the cost-benefit profile of SPE-based sensors, expanding their applications in environmental monitoring, food safety, and clinical diagnostics [102].
Conclusion
Nanomaterial-modified screen-printed electrodes represent a transformative technology for pesticide analysis, successfully bridging the gap between laboratory-based precision and field-deployable convenience. The integration of advanced nanomaterials with SPE platforms has demonstrated remarkable improvements in detection sensitivity, selectivity, and operational stability, enabling rapid screening of pesticide residues at concentrations relevant to regulatory standards. The convergence of multiple detection methodologies—including enzymatic inhibition, immunosensing, and direct electrochemical approaches—provides versatile solutions for diverse analytical scenarios. Future directions should focus on developing multiplexed platforms for simultaneous multi-analyte detection, enhancing sensor longevity for continuous monitoring applications, integrating wireless connectivity for real-time data transmission, and validating these technologies for emerging biomedical applications including exposure biomonitoring and epidemiological studies. As research advances, these portable sensing platforms hold significant potential to revolutionize pesticide monitoring paradigms from agricultural fields to clinical settings, ultimately contributing to enhanced public health protection and personalized exposure assessment.
References
- 1. Development of the screen-printed electrodes: A mini ... [https://www.sciencedirect.com/science/article/pii/S2352186422003455]
- 2. Applications of chemically modified screen-printed electrodes ... [https://pubs.rsc.org/en/content/articlehtml/2024/ra/d4ra02470b]
- 3. Research Advances in Nanosensor for Pesticide Detection in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12298915/]
- 4. Surface Modification of Screen-Printed Carbon Electrodes [https://www.mdpi.com/2079-6412/15/10/1182]
- 5. Fabrication and Evaluation of Screen-Printed Electrodes ... [https://www.mdpi.com/2073-4360/17/15/2088]
- 6. Fabrication of flexible electronics by screen printing with ... [https://www.sciencedirect.com/science/article/pii/S2213846325000677]
- 7. Development of a flow system for decentralized ... [https://pubs.rsc.org/en/content/articlelanding/2024/an/d4an00616j]
- 8. Carbon screen printed electrodes for academic research [https://www.zimmerpeacock.com/2025/06/15/carbon-screen-printed-electrodes-for-academic-research/]
- 9. Recent advances on nanomaterial-modified film-electrode- ... [https://www.sciencedirect.com/science/article/abs/pii/S2451910323002132]
- 10. Electrochemical (Bio)Sensors for Pesticides Detection Using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7236603/]
- 11. Combination of miniature electrode systems via ... [https://www.sciencedirect.com/science/article/abs/pii/S2214158824000278]
- 12. Innovative Carbonaceous Materials and Metal/Metal Oxide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11642939/]
- 13. Applications of chemically modified screen-printed electrodes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11367709/]
- 14. Development of the screen-printed electrodes: A mini ... [https://ui.adsabs.harvard.edu/abs/2022EnvTI..2802922L/abstract]
- 15. A brief review on methods and materials for electrode ... [https://link.springer.com/article/10.1007/s44373-024-00012-8]
- 16. Modification of a pencil graphite electrode with halloysite ... [https://www.nature.com/articles/s41598-025-09917-9]
- 17. Advanced strategies in electrode engineering and ... [https://www.sciencedirect.com/science/article/abs/pii/S2352152X2303551X]
- 18. Protocol for fabricating pseudocapacitor electrodes using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10374872/]
- 19. Reviewing neonicotinoid detection with electroanalytical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11189332/]
- 20. Current electroanalytical approaches in the carbamates ... [https://www.sciencedirect.com/science/article/abs/pii/S0308814623005174]
- 21. Organophosphate - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/physics-and-astronomy/organophosphate]
- 22. A Microbial Electrochemical Technology to Detect and Degrade Organophosphate Pesticides - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8554842/]
- 23. An electrochemical AChE-based biosensor for organophosphate pesticides using a modified CuNWs/rGO nanocomposite on a screen-printed carbon electrode - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0308814623020496]
- 24. Electrochemical sensors of neonicotinoid insecticides ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914023010056]
- 25. Electrochemical (Bio)Sensors for Pesticides Detection ... [https://www.mdpi.com/2079-6374/10/4/32]
- 26. Nanomaterial Labels in Electrochemical Immunosensors ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2819410/]
- 27. Advances in aptamer-based electrochemical biosensors for ... [https://pubs.rsc.org/en/content/articlehtml/2025/nh/d5nh00368g?page=search]
- 28. Nanomaterial-modified electrochemical aptasensors for ... [https://pubs.rsc.org/en/content/articlehtml/2025/an/d5an00097a]
- 29. Aptamer and Oligonucleotide-Based Biosensors for Health ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12109747/]
- 30. From Antibodies to Molecularly Imprinted Polymers (MIPs) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8883926/]
- 31. Molecularly Imprinted Polymers for Biomarker Detection [https://www.sciencedirect.com/science/article/pii/S0165993625003553]
- 32. Recent Advances in Nanomaterial-Based Biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9405907/]
- 33. Nanomaterial-Enhanced Biosensing: Mechanisms and ... [https://pubmed.ncbi.nlm.nih.gov/40167299/]
- 34. Disposable Screen Printed Electrochemical Sensors: Tools for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4118360/]
- 35. Nanoparticle-supported electrochemical sensors for ... [https://www.sciencedirect.com/science/article/pii/S2949771X25000076]
- 36. Recent Advances in Nanomaterial-Based Biosensors for ... [https://www.mdpi.com/2079-6374/12/8/572]
- 37. Gold nanoparticle-modified screen-printed carbon ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11954144/]
- 38. Research progress on nanomaterial-empowered ... [https://pubs.rsc.org/en/content/articlehtml/2025/ra/d5ra04555j?page=search]
- 39. A simple and sensitive approach for the electrochemical ... [https://pubs.rsc.org/en/content/articlelanding/2021/ra/d0ra08723h]
- 40. understanding-electrochemical-detection-principles ... [https://www.transmittershop.com/blog/understanding-electrochemical-detection-principles-techniques-and-environmental-application/]
- 41. Screen-Printed Electrodes: Fabrication, Modification, and ... [https://www.mdpi.com/2227-9040/11/2/113]
- 42. Electrochemical Sensors and Their Applications: A Review [https://www.mdpi.com/2227-9040/10/9/363]
- 43. Highly sensitive capacitance-based nitrite sensing using... [https://pubs.rsc.org/en/content/articlehtml/2023/ra/d3ra03898j]
- 44. An Overview on the Mechanisms and Applications of ... [https://pubmed.ncbi.nlm.nih.gov/32551623/]
- 45. Determination of organophosphate and carbamate ... [https://www.sciencedirect.com/science/article/abs/pii/S0003267004006385]
- 46. Immobilization of Acetylcholinesterase on Screen-Printed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3264471/]
- 47. BVT SPE with immobilized Acetylcholinesterase [https://www.palmsens.com/product/ac1-ache/]
- 48. The Applications of Electrochemical Immunosensors in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10144570/]
- 49. Nanomaterials-Enhanced Multiplex Immunosensing ... [https://portal.nifa.usda.gov/web/crisprojectpages/1015285-nanomaterials-enhanced-multiplex-immunosensing-device-for-rapid-and-sensitive-detection-of-pesticide-residues.html]
- 50. Current Conjugation Methods for Immunosensors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5977292/]
- 51. Development, optimization and validation of an ... [https://www.sciencedirect.com/science/article/pii/S0956713523002591]
- 52. Non-enzymatic electrochemical detection of electro- ... [https://www.sciencedirect.com/science/article/abs/pii/S1385894725024131]
- 53. Non-enzymatic electrochemical sensors based on ... [https://www.sciencedirect.com/science/article/abs/pii/S0924224425000172]
- 54. Bacterial detection with electrochemical, SERS, and ... [https://pubs.rsc.org/en/content/articlehtml/2025/an/d5an00428d]
- 55. Advancements and challenges on SERS-based ... [https://www.sciencedirect.com/science/article/abs/pii/S0924224424003480]
- 56. SERS Tags for Biomedical Detection and Bioimaging - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8825578/]
- 57. Progress in nanomaterials: Synthesis, characterization, ... [https://www.sciencedirect.com/science/article/pii/S2949829525001329]
- 58. Recent Progress in Non-Enzymatic Electroanalytical ... [https://www.mdpi.com/2079-6374/12/5/263]
- 59. Electrochemical sensors: Types, applications, and the ... [https://www.sciencedirect.com/science/article/pii/S0956566324011060]
- 60. Fabrication and characterization of nanomaterial-based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2917878/]
- 61. Nano-fabrication of plasmonic sensors [https://bio-protocol.org/exchange/minidetail?id=1652076&type=30]
- 62. Carbon Nanomaterials-Based Screen-Printed Electrodes for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10136782/]
- 63. Combatting Organophosphate Toxicity: Novel Strategies ... [https://www.sciencedirect.com/science/article/abs/pii/S016599362500319X]
- 64. Promising Electrode Surfaces, Modified with Nanoparticles ... [https://www.mdpi.com/2076-3417/13/9/5391]
- 65. Impact of cleaning procedures on screen-printed gold ... [https://link.springer.com/article/10.1007/s10800-025-02271-8]
- 66. Pesticide residue detection techniques for increasing ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2025.1694779/full]
- 67. A screen-printed electrode modified with gold ... [https://www.sciencedirect.com/science/article/pii/S2405844023025343]
- 68. A universal surface reduction passivation strategy towards ... [https://www.sciopen.com/article/10.26599/NR.2025.94907599]
- 69. Compensate for or Minimize Matrix Effects? Strategies ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7412464/]
- 70. Strategies for the Detection and Elimination of Matrix ... [https://www.chromatographyonline.com/view/strategies-detection-and-elimination-matrix-effects-quantitative-lc-ms-analysis]
- 71. Enhancing Sensitivity and Selectivity: Current Trends in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11505628/]
- 72. Advancements in electrochemiluminescence-based ... [https://www.sciencedirect.com/science/article/pii/S2214180424000904]
- 73. Nanozymes expanding the boundaries of biocatalysis [https://www.nature.com/articles/s41467-025-62063-8]
- 74. Intelligent nanozymes: Biomimetic design, mechanisms ... [https://www.sciencedirect.com/science/article/pii/S2667325824005028]
- 75. Nanozyme-based wearable biosensors for application in ... [https://www.sciencedirect.com/science/article/pii/S2589004225000227]
- 76. Nanozymes-recent development and biomedical applications [https://pmc.ncbi.nlm.nih.gov/articles/PMC8864828/]
- 77. Exploring Nucleic Acid Nanozymes: A New Frontier in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11940440/]
- 78. A microneedle-based integrated three- electrode system for pesticide ... [https://pubs.rsc.org/en/content/articlelanding/2025/an/d5an00430f]
- 79. Design of screen-printed potentiometric platform for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10391324/]
- 80. Innovative electrochemical detection of emerging ... [https://www.sciencedirect.com/science/article/abs/pii/S1572665725005673]
- 81. Voltammetric sensors modified with nanomaterials [https://link.springer.com/article/10.1007/s44373-025-00027-9]
- 82. Immobilization of 4-MBA & Cu2+ on Au nanoparticles ... [https://pubmed.ncbi.nlm.nih.gov/39798418/]
- 83. Nanostructured cadmium sulfide-modified screen-printed ... [https://www.sciencedirect.com/science/article/abs/pii/S0167732224007670]
- 84. An applied potential powered screen-printed electrode for ... [https://www.sciencedirect.com/science/article/abs/pii/S092540052400008X]
- 85. Current Advances in Aptasensors for Pesticide Detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC11930883/]
- 86. Constructing a limit model and developing an HPLC-MS/MS ... [https://pubs.rsc.org/en/content/articlehtml/2025/ra/d4ra08953g]
- 87. Development and Validation of LC–MS/MS and IC–HRMS ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12387874/]
- 88. FDA Reviewer Guidance: Validation of Chromatographic ... [https://www.gmp-compliance.org/guidelines/gmp-guideline/fda-reviewer-guidance-validation-of-chromatographic-methods-1994]
- 89. A comparison of the determination of multiple pesticide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11123614/]
- 90. Next-generation nanomaterials-based biosensors: Real- ... [https://www.sciencedirect.com/science/article/pii/S2589234724004044]
- 91. Nanotechnology-Enabled Biosensors: A Review of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9856107/]
- 92. Advances in Two-Dimensional Nanomaterials for ... [https://link.springer.com/article/10.1007/s11220-025-00686-3]
- 93. Rapid Multi-Residue Detection Methods for Pesticides and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7464912/]
- 94. Disposable electrochemical biosensor based on surface- ... [https://pubs.rsc.org/en/content/articlelanding/2019/ay/c9ay00852g]
- 95. Recent advances in nanomaterials-based electrochemical ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993620302703]
- 96. A Comprehensive Review of Quality Control and Reliability ... [https://www.mdpi.com/2227-7080/13/3/94]
- 97. Toxicity of Carbon Nanomaterials—Towards Reliable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8305450/]
- 98. Surface-enhanced Raman spectroscopy as a tool for food and ... [https://pubs.rsc.org/en/content/articlehtml/2025/na/d5na00539f]
- 99. What are the Main Sensor Methods for Quantifying Pesticides ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6680408/]
- 100. Screen Printed Electrodes Market Size, Share & Growth ... [https://www.researchnester.com/reports/screen-printed-electrodes-market/7275]
- 101. Screen-Printed Electrodes Analysis 2025 and Forecasts ... [https://www.archivemarketresearch.com/reports/screen-printed-electrodes-387891]
- 102. Advancing point-of-care testing with nanomaterials-based ... [https://www.sciencedirect.com/science/article/pii/S2666351125000038]
- 103. Disposable Printed Electrode Made with Chinese Shellac ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12371578/]