Smartphone-Integrated Biosensors for Visual Pesticide Detection: A New Paradigm for On-Site Food Safety and Environmental Monitoring
This article provides a comprehensive analysis of the latest advancements in smartphone-integrated biosensors specifically designed for the visual detection of pesticide residues.
Smartphone-Integrated Biosensors for Visual Pesticide Detection: A New Paradigm for On-Site Food Safety and Environmental Monitoring
Abstract
This article provides a comprehensive analysis of the latest advancements in smartphone-integrated biosensors specifically designed for the visual detection of pesticide residues. Tailored for researchers, scientists, and drug development professionals, it explores the foundational principles of these portable analytical platforms, delves into novel methodologies such as ratiometric fluorescent probes and molecularly imprinted polymers (MIPs), and addresses critical challenges in sensor calibration and real-world deployment. The scope extends to rigorous performance validation against traditional chromatographic methods and discusses the transformative potential of integrating artificial intelligence (AI) and IoT connectivity for enhancing diagnostic accuracy and enabling widespread, decentralized monitoring in agricultural, clinical, and environmental contexts.
The Science Behind Smartphone Biosensors: Principles, Components, and Recognition Elements
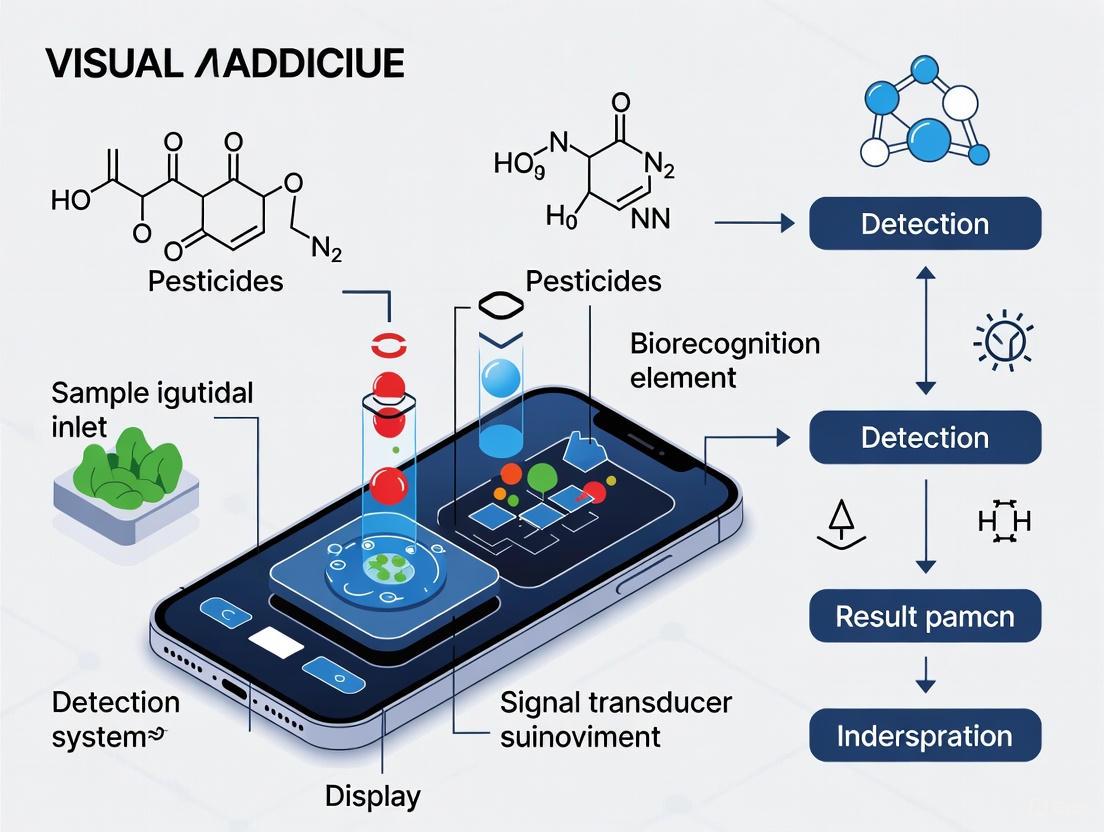
Smartphone-integrated biosensors represent a transformative approach for the on-site detection of pesticides, merging the specificity of biological recognition with the ubiquity and processing power of mobile devices. These systems are particularly valuable for environmental and food safety monitoring, such as detecting organophosphorus and carbamate pesticides in tea and other agricultural products [1]. The core working principle involves a sequential process: a biorecognition element first selectively binds to the target pesticide, this binding event is transduced into a measurable optical signal, and the smartphone then captures and processes this signal to provide a quantitative readout [2] [3] [4]. This document details the application notes and experimental protocols underlying this technology, providing a framework for researchers and scientists engaged in its development and application.
Core Principles and Biosensor Architecture
The operation of a smartphone-integrated biosensor for visual pesticide detection rests on three foundational pillars: biorecognition, signal transduction, and smartphone-based readout.
Biorecognition Elements
The specificity of the biosensor is determined by its biorecognition element, which selectively interacts with the target analyte. Common types include:
- Enzymes: Acetylcholinesterase (AChE) is widely used for detecting organophosphorus and carbamate pesticides. These pesticides inhibit AChE activity, reducing the rate of enzymatic hydrolysis of its substrate (e.g., acetylthiocholine). The subsequent reduction in a colored or electroactive product formation serves as the basis for detection [1] [4].
- Antibodies: These proteins offer high affinity and specificity for a wide range of pesticide targets. Immunosensors utilize the antibody-antigen binding event, which can be detected through label-based or label-free methods [4].
- Nucleic Acid Aptamers: Single-stranded DNA or RNA molecules that fold into specific three-dimensional structures to bind target molecules with antibody-like affinity. They are synthetic, stable, and easily modified, making them emerging tools for biosensing [4].
- Biomimetic Receptors: This category includes Molecularly Imprinted Polymers (MIPs), which are artificial receptors containing tailor-made binding sites complementary to the target pesticide. They are prized for their high stability and cost-effectiveness [4].
Signal Transduction and Transduction
Following biorecognition, the binding event must be converted into a quantifiable signal. For visual detection, optical transduction is paramount:
- Colorimetric Transduction: The most common method for smartphone-based visual detection. It often involves nanoparticles, such as gold nanoparticles (AuNPs), which exhibit intense colors due to surface plasmon resonance. A reaction between the bioreceptor and the target analyte can induce a color change or a shift in the absorption spectrum of the AuNPs [1] [5]. For instance, AChE inhibition can be coupled to a reaction that prevents the aggregation of AuNPs, resulting in a distinct color change from red to blue [6].
- Fluorescence Transduction: Some biosensors use fluorescent dyes or materials (e.g., quantum dots, metal-organic frameworks). The presence of the pesticide may quench or enhance the fluorescence intensity, which can be captured by the smartphone camera [1] [5].
Smartphone Readout and Data Processing
The smartphone serves as a portable spectrophotometer and data processor [2] [3]. The core steps are:
- Image Acquisition: The smartphone camera captures an image of the colorimetric reaction under controlled lighting conditions, often using a custom attachment to ensure consistency [6].
- Image Pre-processing: Algorithms correct for variable lighting conditions (e.g., using the gray world algorithm or a reference color card), segment the region of interest (the test strip), and extract color values (typically in RGB, HSV, or LAB color spaces) [6].
- Data Analysis and Quantification: Machine learning models, such as Support Vector Machines (SVM) for classification and Support Vector Regression (SVR) for continuous concentration prediction, are deployed on the smartphone to map the extracted color features to pesticide concentration [6]. This process mitigates the subjectivity of visual interpretation and enhances accuracy.
The following diagram illustrates the complete integrated workflow from sample to result.
Performance Comparison of Biosensing Technologies
The analytical performance of biosensors varies significantly based on the detection technique and the biorecognition element employed. The table below summarizes key performance metrics for common biosensor types used in pesticide detection, compared to traditional methods.
Table 1: Comparative Analysis of Biosensor Performance for Pesticide Detection
| Detection Technique | Biorecognition Element | Typical Detection Limit | Assay Time | Key Advantages | Primary Limitations |
|---|---|---|---|---|---|
| Electrochemical [1] [3] | Enzymes (AChE), Antibodies | nM - pM | 5 - 30 min | High sensitivity, portability, cost-effectiveness | Signal drift, electrode fouling |
| Fluorescence [1] [5] | Aptamers, Antibodies | pM range (MOF-enhanced) | 15 - 60 min | Very high sensitivity, multiplexing potential | Requires light source, can be complex |
| Colorimetric (Smartphone) [1] [6] | Enzymes (AChE), Antibodies | nM - µM | 10 - 30 min | Simplicity, true portability, low cost | Susceptible to ambient light interference |
| Surface Plasmon Resonance (SPR) [1] | Antibodies | nM range | 10 - 20 min | Label-free, real-time monitoring | Expensive instrumentation, bulky |
| Chromatography (GC/HPLC) [1] | N/A | nM - pM | Hours | Gold standard, high accuracy & precision | Lab-bound, expensive, requires trained personnel |
Experimental Protocol: Smartphone-Based Colorimetric Detection of Pesticides using an Acetylcholinesterase (AChE) Biosensor
This protocol provides a detailed methodology for detecting organophosphorus pesticides using an AChE-inhibited reaction on a test strip, with quantification via a smartphone application.
Principle
The assay is based on the inhibition of AChE. In the absence of pesticide, AChE hydrolyzes acetylthiocholine to thiocholine, which reduces a chromogen (e.g., Ellman's reagent) to produce a yellow color. When pesticides are present, they inhibit AChE, reducing the generation of thiocholine and resulting in a diminished color intensity. This color change is inversely proportional to the pesticide concentration and is quantified by a smartphone app [1] [6].
Materials and Equipment
- Smartphone with custom-built analysis application (e.g., built with Flutter framework) [6].
- Test strips impregnated with AChE and substrate/chromogen system [6].
- Gold Nanoparticles (AuNPs): Can be used as colorimetric probes in alternative assay designs [6].
- Reference color card (for illumination correction).
- Sample solutions: Pesticide standards and unknown samples.
- Pipettes and microcentrifuge tubes.
Procedure
Sample Preparation:
- Prepare a series of pesticide standard solutions in a suitable buffer (e.g, phosphate buffer saline) to generate a calibration curve.
- Prepare unknown samples by extracting and filtering, if necessary.
Assay Execution:
- Apply a fixed volume (e.g., 50 µL) of standard or sample solution onto the test strip.
- Incubate the test strip at room temperature for a defined period (e.g., 10-15 minutes) to allow for the enzyme inhibition reaction to complete.
- After incubation, initiate the color development reaction if required by the specific test strip design.
Image Acquisition:
- Place the developed test strip and the reference color card in a well-lit area, avoiding shadows and glare.
- Open the smartphone application. Use the in-app guide to align the test strip and color card within the camera's field of view.
- Ensure stable exposure and capture the image.
Data Analysis:
- The application will automatically pre-process the image: correct illumination using the reference card, and segment the region of interest on the test strip.
- The application will extract color features (e.g., RGB values) from the segmented area.
- A pre-trained machine learning model (SVM/SVR) will analyze the color features and output the predicted pesticide concentration [6].
The specific data processing workflow within the smartphone is detailed below.
Data Interpretation
- The application provides a numerical value of the pesticide concentration.
- Results can be categorized into "safe," "warning," or "hazardous" based on predefined regulatory thresholds (e.g., from GB 2763-2021 or EU MRLs) [1].
- Data, along with timestamp and geolocation, can be optionally uploaded to a cloud database for large-scale environmental monitoring [6].
The Scientist's Toolkit: Essential Research Reagent Solutions
The development and execution of smartphone-based biosensors rely on a core set of reagents and materials. The following table catalogues key components and their functions.
Table 2: Key Research Reagents and Materials for Biosensor Development
| Item | Function/Application | Key Characteristics |
|---|---|---|
| Acetylcholinesterase (AChE) [1] [4] | Primary biorecognition element for organophosphorus/carbamate pesticides. Enzyme inhibition is the basis of detection. | High specific activity, purity, and stability. |
| Polyclonal/Monoclonal Antibodies [4] | Biorecognition element for specific pesticide targets in immunosensors. | High affinity and specificity. Monoclonal offers better reproducibility. |
| Nucleic Acid Aptamers [4] | Synthetic biorecognition element; can be selected for various targets via SELEX. | High thermal stability, easily synthesized and modified. |
| Gold Nanoparticles (AuNPs) [5] [6] | Colorimetric signal probe; color changes upon aggregation or interaction with analyte. | High extinction coefficient, tunable surface chemistry. |
| Molecularly Imprinted Polymers (MIPs) [4] | Biomimetic synthetic receptor; template-shaped cavities for pesticide binding. | High chemical/thermal stability, low-cost production. |
| Metal-Organic Frameworks (MOFs) [1] [5] | Fluorescence signal amplification; can be used to enhance sensor sensitivity. | High porosity, tunable structure, and strong luminescence. |
Key Components of a Smartphone-Integrated Biosensing System
Smartphone-integrated biosensing systems represent a convergence of specific biological recognition elements and the versatile data processing, connectivity, and imaging capabilities of modern smartphones. These systems are primarily designed for point-of-care (POC) and point-of-need testing, enabling the decentralized detection of analytes such as pesticide residues in food and environmental samples [7] [8]. Their architecture is defined by the location of the biosensing function (on- or off-phone) and the locus of data processing (local on the smartphone or remotely on a server) [7]. For visual pesticide detection, systems leveraging the smartphone's built-in camera for optical readout are particularly prominent, functioning as portable spectrophotometers or fluorimeters [9]. This document outlines the key components, performance metrics, and detailed experimental protocols for assembling and utilizing such systems, with a specific focus on applications in food safety and environmental monitoring.
Core System Components and Their Functions
A fully functional smartphone-integrated biosensor comprises several integrated subsystems: the biological recognition element, the transducer, the smartphone with its hardware and software, and, for some configurations, external accessories and data servers.
Table 1: Key Components of a Smartphone-Integrated Biosensing System
| Component Category | Specific Element | Function & Description |
|---|---|---|
| Biological Recognition | Acetylcholinesterase (AChE) | Enzyme inhibited by Organophosphorus (OP) pesticides; basis for enzymatic biosensors [10]. |
| Alkaline Phosphatase (ALP) | Enzyme used in enzyme-linked assays; its inhibition can be correlated to pesticide concentration [11]. | |
| Antibodies & Aptamers | Provide high specificity for immunoassays and aptamer-based sensors [1] [9]. | |
| Transducer & Signal Conversion | Polyaniline Nanofibers (PAnNFs) | Conducting polymer; conductance changes with proton doping during ACh hydrolysis, enabling resistive sensing [10]. |
| Carbon Nanotubes (CNTs) | Nanomaterial used in nanocomposite films to enhance conductivity and sensor performance [10]. | |
| Fluorescent Markers (e.g., DFQ) | Molecule produced in enzymatic reactions; its fluorescence intensity, when excited, is measured quantitatively [11]. | |
| Smartphone Hardware & Software | CMOS Camera | Acts as a optical detector for colorimetric, fluorescence, or label-free assays [7] [9]. |
| Mobile Application (App) | Controls data acquisition, processing, analysis, visualization, and sharing of results [10] [8]. | |
| CPU/Connectivity (Bluetooth, USB) | Provides processing power and a link to external sensors or cloud servers for data handling [7] [12]. | |
| External Accessories | Portable Fluorescence Device | Custom attachment with LEDs and filters to create a controlled environment for fluorescence excitation and emission [11]. |
| Cradle Attachment with Diffraction Grating | Converts the smartphone camera into a spectrometer for wavelength-specific measurements [9]. | |
| Microfluidic Paper-Based Device (μPAD) | Provides a low-cost, disposable platform with hydrophilic/hydrophobic channels to conduct assays [3]. |
Performance Metrics of Representative Systems
The analytical performance of developed systems is critical for assessing their applicability. The following table summarizes data from recent research on smartphone-based biosensors for pesticide detection.
Table 2: Analytical Performance of Smartphone-Based Biosensors for Pesticide Detection
| Detection Principle | Target Analyte | Linear Range | Limit of Detection (LOD) | Real-Sample Application | Citation Source |
|---|---|---|---|---|---|
| Fluorescence Biosensor (ALP-based) | Malathion (Organophosphorus) | 0.1 - 1 ppm | 0.05 ppm | Vegetable samples | [11] |
| Resistive Biosensor (AChE/PAnNF/CNT) | Paraoxon-Methyl (Organophosphate) | 1 ppt - 100 ppb | 0.304 ppt | Food and environmental water | [10] |
| Electrochemical Biosensor (General) | Various Pesticides | Not Specified (High Sensitivity) | nM to pM levels | Tea leaves | [1] |
Detailed Experimental Protocols
Protocol 1: Fluorescence-Based Detection of Organophosphorus Pesticides
This protocol is adapted from a study detailing a smartphone-based fluorescence biosensor for malathion [11].
I. Research Reagent Solutions
- L-Ascorbic Acid 2-phosphate sesquimagnesium salt hydrate (AAP) Solution: Serves as the enzyme substrate.
- Alkaline Phosphatase (ALP) Enzyme Solution: The enzyme whose activity is inhibited by the target pesticide.
- o-Phenylenediamine (OPD) Solution: Reacts with ascorbic acid to form the fluorescent product DFQ.
- Organophosphorus Pesticide Standard Solutions: (e.g., Malathion) in a range of known concentrations for calibration.
- Sample Extraction Buffer: Suitable for extracting pesticides from vegetable matrices.
II. Procedure
- Sample Preparation: Homogenize the vegetable sample (e.g., 1 g) and extract the analyte using a suitable buffer. Centrifuge and filter the supernatant to obtain a clear test solution.
- Reaction Incubation: In a microcentrifuge tube, mix the following:
- A fixed volume of the sample extract or pesticide standard.
- A known activity of ALP enzyme solution.
- Incubate the mixture for 10 minutes at 37°C to allow for potential enzyme inhibition by the pesticide.
- Fluorescent Product Generation: To the same tube, add:
- AAP substrate solution.
- OPD solution.
- Incubate for another 20 minutes at 37°C. In the absence of inhibitor, ALP converts AAP to ascorbic acid, which then reacts with OPD to yield the fluorescent molecule DFQ. The presence of pesticide inhibits ALP, reducing DFQ formation proportionally.
- Smartphone Measurement:
- Transfer the reaction solution to a cuvette or a well in a microtiter plate.
- Place the container into the portable fluorescence device attachment, which is equipped with an appropriate excitation LED.
- Use the smartphone app to capture an image of the fluorescence emission or to record the RGB values of the solution.
- Data Analysis: The smartphone app converts the fluorescence intensity (via RGB values) into a concentration. A calibration curve, generated from standards of known concentration, is used for interpolation.
Protocol 2: Resistive Biosensor for Organophosphate Pesticides
This protocol is based on an integrated smartphone/resistive biosensor for sensitive OP pesticide monitoring [10].
I. Research Reagent Solutions
- Acetylcholinesterase (AChE) Enzyme Solution: The biological recognition element.
- Polyaniline Nanofibers (PAnNFs) Suspension: The conductive transducer material.
- Carbon Nanotube (CNT) Suspension: Used to form a conductive nanocomposite.
- Chitosan Solution: A biopolymer for forming a stable nanocomposite film.
- Acetylcholine (ACh) Substrate Solution: The hydrolyzed substrate.
- Pesticide Standard Solutions: (e.g., Paraoxon-Methyl) for calibration.
II. Procedure
- Biosensor Fabrication:
- Prepare a nanocomposite by mixing AChE, PAnNFs, CNTs, and chitosan in a defined ratio.
- Deposit a small volume of the nanocomposite onto a gold interdigitated electrode (IDE).
- Allow the film to dry and crosslink, forming the functional resistive biosensor.
- Baseline Measurement:
- Connect the IDE to a readout circuit that can communicate with the smartphone (e.g., via USB or Bluetooth).
- Using the smartphone app, measure the initial conductance of the biosensor film.
- Inhibition and Measurement:
- Expose the biosensor to a sample solution (extracted from food or water) containing the target pesticide for a fixed time (e.g., 10 minutes).
- Wash the sensor gently to remove unbound molecules.
- Introduce the substrate, acetylcholine (ACh), to the sensor.
- The smartphone app records the change in conductance over time. In a pesticide-free sample, AChE hydrolyzes ACh, releasing protons that dope the PAnNFs and cause a large conductance increase. If AChE is inhibited by pesticides, the conductance change is proportionally smaller.
- Data Processing: The smartphone app analyzes the rate of conductance change or its absolute value. The degree of inhibition is calculated relative to a negative control and correlated to the pesticide concentration via a pre-loaded calibration curve.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents for Smartphone-Based Pesticide Biosensors
| Reagent/Material | Function in the Experiment |
|---|---|
| Acetylcholinesterase (AChE) | Core biorecognition element for OP pesticides; its inhibition is the basis for quantification in enzymatic sensors [10]. |
| Alkaline Phosphatase (ALP) | Enzyme used in alternative enzyme-inhibition assays; its activity is modulated by the presence of inhibitors [11]. |
| Polyaniline Nanofibers (PAnNFs) | Conducting polymer transducer; its electronic properties (conductance) change in response to biochemical reactions (proton doping) [10]. |
| Carbon Nanotubes (CNTs) | Nanomaterial used to enhance electron transfer and improve the sensitivity and stability of electrochemical/resistive biosensors [10]. |
| L-Ascorbic Acid 2-phosphate (AAP) | Enzyme substrate that is converted by ALP to ascorbic acid, a key reactant in a subsequent fluorescence-generating reaction [11]. |
| o-Phenylenediamine (OPD) | Chemical compound that reacts with ascorbic acid to produce a fluorescent product (DFQ), enabling optical detection [11]. |
| Gold Interdigitated Electrodes (IDEs) | The physical transducer platform where the biosensing film is immobilized and electrical (resistive) measurements are taken [10]. |
| Specific Antibodies/Aptamers | High-affinity recognition elements for designing immunosensors or aptasensors with high specificity for target pesticide molecules [1] [9]. |
The development of robust, selective, and sensitive biosensors hinges on the performance of their molecular recognition elements. These components are responsible for the specific binding and identification of target analytes within complex sample matrices. Within the specific context of developing smartphone-integrated biosensors for the visual detection of pesticides, the choice of recognition element dictates the sensor's overall applicability, sensitivity, and potential for field deployment. This document provides detailed application notes and protocols for four primary classes of advanced recognition elements—Enzymes, Antibodies, Aptamers, and Molecularly Imprinted Polymers (MIPs)—framed within the demands of modern, portable biosensing platforms. The convergence of these elements with smartphone-based detection heralds a new era of on-site analysis, enabling rapid and quantitative monitoring of pesticide residues for environmental and food safety [1].
Performance Comparison of Advanced Recognition Elements
The selection of an appropriate recognition element requires a balanced consideration of its inherent properties. The following table summarizes the key characteristics of each element type, providing a guide for selection based on the requirements of a specific biosensing application, particularly for pesticide detection.
Table 1: Comparative analysis of advanced recognition elements for biosensing.
| Recognition Element | Affinity & Specificity | Stability & Production | Key Advantages | Primary Limitations | Common Transduction Methods |
|---|---|---|---|---|---|
| Enzymes | Moderate; specificity for substrate catalysis | Low thermal/operational stability; complex purification | Natural catalytic activity; signal amplification | Susceptible to inhibition; limited target scope | Electrochemical, Optical (Colorimetric, Fluorescent) |
| Antibodies | High (pM-nM); high specificity for a single epitope | Moderate stability; sensitive to conditions; requires animal hosts | Well-established, commercial availability; high specificity | Batch-to-batch variation; expensive production; animal use | ELISA (Colorimetric, Chemiluminescent), SPR, Fluorescent |
| Aptamers | High (nM-pM); high specificity for small molecules | High thermal/chemical stability; chemical synthesis | Small size; tunable affinity; label-free detection possible | Susceptible to nuclease degradation; complex SELEX process | Fluorescent (FRET), Electrochemical, SPR, Colorimetric |
| Molecularly Imprinted Polymers (MIPs) | Moderate to High; specificity mimics antibodies | Excellent stability (thermal, pH, solvent); chemical synthesis | Robustness; low-cost; reusability; long shelf-life | Occasional heterogeneity in binding sites | Electrochemical, Optical, SPR |
Detailed Element Analysis and Protocols
Antibodies and the Enzyme-Linked Immunosorbent Assay (ELISA)
Antibodies are immunoglobulins that bind to specific molecular epitopes with high affinity. The sandwich ELISA is a premier format for achieving high sensitivity and specificity, making it a gold standard for protein detection [13] [14]. In this format, a capture antibody is immobilized on a surface to bind the target antigen from a sample, after which a second, enzyme-conjugated detection antibody is added to complete the "sandwich." The enzyme, such as Horseradish Peroxidase (HRP), then catalyzes the conversion of a substrate into a colored, fluorescent, or chemiluminescent product, enabling quantification [15].
Table 2: Key research reagents for antibody-based biosensing.
| Reagent / Solution | Function in the Protocol |
|---|---|
| Capture Antibody | Immobilized on the microplate to specifically bind the target pesticide or its derivative. |
| Enzyme-Conjugated Detection Antibody | Binds a different epitope on the captured target and provides the signal via enzyme catalysis. |
| Blocking Buffer (e.g., BSA or Skim Milk) | Blocks unsaturated binding sites on the microplate to minimize non-specific adsorption. |
| Coating Buffer (e.g., Carbonate-Bicarbonate, pH 9.4) | Provides optimal pH and ionic conditions for passive adsorption of the capture antibody to the plate. |
| Enzyme Substrate (e.g., ABTS for HRP) | Converted by the enzyme into a measurable product, generating the detection signal. |
Protocol: Sandwich ELISA for Pesticide Detection
- Coating: Dilute the capture antibody in a carbonate-bicarbonate coating buffer (pH 9.4) to a concentration of 2–10 µg/mL. Add 100 µL per well to a 96-well microplate and incubate for 16 hours at 4°C [15].
- Blocking: Wash the plate three times with PBS containing 0.1% Tween 20 (wash buffer). Add 200 µL of blocking buffer (e.g., 5% skim milk or BSA in PBS) to each well and incubate for 1–2 hours at room temperature. Wash three times.
- Sample Incubation: Add 100 µL of the sample or pesticide standard to each well. Incubate for 1–2 hours at 37°C to allow antigen binding. Wash thoroughly to remove unbound material.
- Detection Antibody Incubation: Add 100 µL of the enzyme-conjugated detection antibody (diluted in blocking buffer) to each well. Incubate for 1–2 hours at 37°C. Wash extensively.
- Signal Development & Smartphone Readout: Add 100 µL of the appropriate substrate solution (e.g., ABTS for HRP). Incubate in the dark for 15–30 minutes. Terminate the reaction if necessary. Instead of a plate reader, place the plate on a uniform light source and capture an image using a smartphone. Analyze the color intensity of each well using image processing software (e.g., ImageJ) to generate a quantitative standard curve and determine analyte concentration [1].
Aptamers and Fluorescent Biosensing
Aptamers are single-stranded DNA or RNA oligonucleotides selected in vitro to bind specific targets with high affinity and specificity, earning them the moniker "chemical antibodies" [16] [17]. Their utility in biosensors is extensive, with fluorescent aptasensors being particularly suitable for integration with smartphone optics. A common mechanism involves a "signal-on" configuration based on Fluorescence Resonance Energy Transfer (FRET), where an aptamer is labeled with a fluorophore whose emission is quenched by a nearby nanomaterial (e.g., graphene oxide) or quencher. Upon binding the target, the aptamer undergoes a conformational change, separating the fluorophore from the quencher and restoring fluorescence [16].
Table 3: Key research reagents for aptamer-based biosensing.
| Reagent / Solution | Function in the Protocol |
|---|---|
| Fluorophore-Labeled Aptamer | The core recognition element; its target-induced conformational change modulates the fluorescence signal. |
| Quencher or Nanomaterial (e.g., Graphene Oxide) | Initially quenches the fluorophore's emission; signal is generated upon displacement. |
| Binding Buffer | Provides optimal ionic strength and pH to facilitate correct aptamer folding and target binding. |
| Gold Nanoparticles (AuNPs) | Used for signal amplification and enhancement in various optical and electrochemical sensors [17]. |
Protocol: 'Signal-On' Fluorescent Aptasensor for Pesticide Detection
- Aptamer Preparation: Reconstitute the fluorophore-labeled aptamer (e.g., specific for a pesticide like ochratoxin A) in the appropriate binding buffer. Anneal the aptamer by heating and slowly cooling to ensure proper folding [16].
- Sensor Assembly & Quenching: Incubate the folded aptamer with the quencher (e.g., graphene oxide) for a fixed time (e.g., 30 minutes) to allow adsorption and fluorescence quenching.
- Target Binding & Signal Recovery: Introduce the sample containing the target pesticide to the aptamer-quencher mixture. Incubate for 30-60 minutes. Target binding will cause the aptamer to change conformation, releasing the fluorophore from the quencher and restoring fluorescence.
- Smartphone Fluorescence Detection: Transfer the solution to a low-volume cuvette or a microfluidic chip designed for smartphone attachment. Using a simple smartphone adapter equipped with a complementary filter set, illuminate the sample with the excitation wavelength and capture the emitted fluorescence. The intensity of the green fluorescence is proportional to the pesticide concentration and can be quantified using a smartphone application [16] [1].
Molecularly Imprinted Polymers (MIPs)
MIPs are synthetic polymers that possess tailor-made recognition sites complementary to a target molecule in shape, size, and functional groups. They are fabricated by polymerizing functional monomers around a template molecule (the target analyte). Subsequent removal of the template leaves behind cavities that exhibit high specificity for the original molecule, functioning as artificial antibodies [18]. Their exceptional physical and chemical stability makes them ideal for harsh environments and reusable sensors.
Table 4: Key research reagents for MIP-based biosensing.
| Reagent / Solution | Function in the Protocol |
|---|---|
| Template Molecule (Target Pesticide) | Serves as the mold around which the complementary cavity is formed during polymerization. |
| Functional Monomer | Contains functional groups that form reversible interactions with the template. |
| Cross-linker | Creates a rigid polymer network that stabilizes the imprinted cavities after template removal. |
| Electrochemical Probe (e.g., [Fe(CN)₆]³⁻/⁴⁻) | Used in electrochemical MIP sensors; its signal is perturbed upon target rebinding. |
Protocol: Electrochemical MIP Nano-sensor for Pesticide Detection
- MIP Synthesis on Electrode: Mix the target pesticide (template), functional monomer (e.g., acrylic acid), and cross-linker (e.g., ethylene glycol dimethacrylate) in a solvent. Add an initiator and deposit the pre-polymerization mixture onto the surface of a working electrode (e.g., glassy carbon or screen-printed electrode). Polymerize via UV irradiation or thermal initiation to form a thin polymer film [18].
- Template Removal: Carefully wash the polymer-coated electrode with a solvent mixture (e.g., methanol:acetic acid) to extract the template molecules from the polymer matrix, leaving behind specific recognition sites.
- Rebinding and Measurement: Incubate the MIP-modified electrode in the sample solution containing the pesticide. The target molecules will selectively rebind to the imprinted cavities. For electrochemical detection, monitor the current of a redox probe (e.g., ferricyanide) using differential pulse voltammetry (DPV) or electrochemical impedance spectroscopy (EIS). The binding of the pesticide hinders electron transfer, leading to a measurable change in current, which is proportional to the pesticide concentration [18].
- Smartphone Integration: Connect the electrochemical sensor to a miniaturized, smartphone-operated potentiostat. The smartphone can control the measurement parameters and record the electrochemical signal. The data is processed by a dedicated app to provide a quantitative result on-screen, enabling fully portable electrochemical detection [18] [1].
The integration of advanced recognition elements with smartphone-based detection platforms creates powerful tools for on-site pesticide monitoring. Antibodies offer proven sensitivity, aptamers provide versatility and stability, and MIPs deliver unmatched robustness and low-cost potential. The choice of element is application-dependent, but the ongoing trend is toward the development of MIPs and aptamers that match the affinity of antibodies while offering superior stability for field use. The future of this field lies in the fusion of these elements with nanomaterials for signal enhancement, microfluidics for automated sample handling, and IoT platforms for real-time data geolocation and sharing, ultimately creating a connected network for environmental and food safety surveillance [18] [1].
Biosensors are analytical devices that combine a biological recognition element with a transducer to produce a measurable signal proportional to the concentration of a target analyte. The transduction mechanism is a fundamental component that defines the sensor's characteristics, performance, and suitability for specific applications. In the context of developing smartphone-integrated biosensors for visual pesticide detection, the choice between optical and electrochemical transduction is particularly critical. This application note provides a comparative overview of these two dominant transduction mechanisms, focusing on their operational principles, performance parameters, and implementation protocols for pesticide detection applications. The integration of these biosensing platforms with smartphone technology represents a frontier in point-of-care testing, enabling rapid on-site analysis for environmental monitoring and food safety.
Fundamental Principles and Mechanisms
Optical Transduction Mechanisms
Optical biosensors detect targets by recognizing changes in optical properties and converting them into readable signals [19]. These sensors employ various optical phenomena including fluorescence, colorimetry, surface plasmon resonance (SPR), and surface-enhanced Raman spectroscopy (SERS) [19]. For pesticide detection, the enzyme inhibition principle is commonly employed, where organophosphorus pesticides inhibit acetylcholinesterase activity, leading to measurable changes in optical signals [20].
Fluorescence-based sensing operates on principles such as Förster Resonance Energy Transfer, where energy transfer occurs between a donor fluorophore and an acceptor quencher. Target-induced conformational changes alter donor-quencher proximity, terminating FRET and restoring fluorescence [19]. Nanomaterials like graphene oxide have been extensively utilized in FRET-based aptasensors due to exceptional photoelectric properties that enable fluorescence quenching [19].
Colorimetric sensing detects color changes visible to the naked eye or through smartphone cameras. Nanozyme-based colorimetric strategies have gained prominence, where nanomaterials with enzyme-like activity catalyze chromogenic reactions [21] [20]. For instance, hydrogen-bonded organic framework nanozymes with peroxidase-like activity can catalyze the oxidation of 3,3',5,5'-tetramethylbenzidine, producing a color change measurable via smartphone [20].
Surface-enhanced Raman scattering provides fingerprint molecular identification through significant enhancement of Raman signals when analytes are adsorbed on nanostructured metal surfaces, enabling highly sensitive detection [21].
Electrochemical Transduction Mechanisms
Electrochemical biosensors measure electrical signals resulting from biochemical interactions at the electrode-solution interface [22]. These sensors encompass techniques including voltammetry, amperometry, potentiometry, electrochemical impedance spectroscopy, and electrochemiluminescence [22].
In amperometric sensors, current is measured at a constant potential applied to the working electrode, with the magnitude proportional to analyte concentration [23]. For organophosphorus pesticide detection, this typically involves measuring changes in cholinesterase activity through substrate hydrolysis [23].
Voltammetric techniques apply a potential sweep and measure resulting current, providing information about redox reactions. The strategic design of electrode surfaces with nanomaterials enhances electron transfer characteristics and loading efficacy of biorecognition elements [22].
Impedimetric sensors monitor changes in electrical impedance resulting from binding events at modified electrode surfaces, often enabling label-free detection [22].
Electrochemiluminescence combines electrochemical and optical methods, whereelectrochemical reactions generate luminescent species, offering high sensitivity with low background signals [22].
Comparative Performance Analysis
The table below summarizes key performance characteristics of optical and electrochemical transduction mechanisms for biosensing applications, particularly focused on pesticide detection.
Table 1: Performance Comparison of Optical and Electrochemical Transduction Mechanisms
| Parameter | Optical Transduction | Electrochemical Transduction |
|---|---|---|
| Sensitivity | High (e.g., LOD of 3.04 ng/mL for chlorpyrifos using HOF nanozyme) [20] | High (e.g., detection limits of 0.11 U/mL for AChE) [23] |
| Selectivity | High (molecular recognition via enzymes, antibodies, aptamers) [19] [20] | High (bioreceptor specificity combined with electrochemical selectivity) [22] |
| Multiplexing Capability | Moderate to high (multiple wavelengths, spatial resolution) [24] | Moderate (multiple electrode arrays, different potentials) [22] |
| Sample Volume | Microliter to milliliter range [20] | Microliter range (miniaturized electrochemical cells) [22] |
| Detection Time | Seconds to minutes (rapid color development) [20] | Seconds to minutes (rapid electron transfer) [23] [22] |
| Instrumentation Complexity | Moderate to high (light sources, detectors) [21] | Low to moderate (potentiostats, readout circuits) [22] |
| Cost | Moderate to high (optical components) [21] | Low (miniaturized electronics) [22] |
| Smartphone Integration | Excellent (built-in cameras for colorimetric/fluorescent detection) [23] [20] | Good (requires external interface circuitry) [23] |
| Reproducibility | Moderate (nanomaterial batch variations) [20] | Moderate to high (electrode surface reproducibility challenges) [22] |
Experimental Protocols
Protocol for Optical Biosensor (Colorimetric Nanozyme-based Detection)
This protocol describes the development of a smartphone-integrated colorimetric biosensor for organophosphorus pesticide detection using HOF nanozymes [20].
Materials and Reagents:
- Acetylcholinesterase (220 U/mg)
- 6,6',6'',6'''-(Pyrene-1,3,6,8-tetrayl) tetrakis(2-naphthoic acid) (H4PTTNA)
- Acetylthiocholine iodide
- 3,3',5,5'-Tetramethylbenzidine
- Hemin
- Sodium alginate
- Calcium chloride
- Organophosphorus pesticide standards (chlorpyrifos, acephate, fenthion)
- Hydrogen peroxide
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
Procedure:
Synthesis of Hemin@HOF Nanozyme:
- Dissolve H4PTTNA (5 mg) in DMSO (1 mL) by sonication for 10 minutes
- Add bovine serum albumin (50 mg) to phosphate buffer (10 mL, 10 mM, pH 7.4)
- Mix H4PTTNA solution with BSA solution under vigorous stirring
- Add Hemin solution (2 mg/mL in DMSO) dropwise to the mixture
- Incubate at room temperature for 12 hours with continuous stirring
- Centrifuge at 12,000 rpm for 15 minutes and wash three times with deionized water
- Resuspend in PBS and store at 4°C until use
Hydrogel Biosensor Preparation:
- Prepare sodium alginate solution (3% w/v) in deionized water
- Mix Hemin@HOF nanozyme suspension with sodium alginate solution at 1:4 volume ratio
- Add AChE enzyme (0.5 U/mL final concentration) to the mixture
- Dropwise add the mixture into CaCl₂ solution (5% w/v) to form hydrogel beads
- Incubate for 30 minutes to complete cross-linking
- Wash hydrogel beads with PBS and store at 4°C in moist conditions
Pesticide Detection Assay:
- Place hydrogel biosensor in microcentrifuge tube
- Add pesticide sample (100 μL) and incubate for 15 minutes at 37°C
- Add ATCh substrate (200 μL, 5 mM) and incubate for 10 minutes
- Add TMB solution (200 μL, 2 mM) and H₂O₂ (50 μL, 10 mM)
- Incubate for 5 minutes to allow color development
- Capture image using smartphone camera under controlled lighting
- Analyze RGB values using color analysis application
Data Analysis:
- Measure blue channel intensity from smartphone images
- Plot intensity versus pesticide concentration for quantification
- Calculate limit of detection using 3σ/slope method
Protocol for Electrochemical Biosensor (Resistive Nanosensor Platform)
This protocol describes the development of a smartphone-integrated resistive nanosensor for organophosphorus pesticide detection via cholinesterase activity monitoring [23].
Materials and Reagents:
- Multiwalled carbon nanotubes
- Polyaniline nanofibers
- Chitosan solution (1% in acetic acid)
- Gold interdigitated electrodes
- Acetylcholinesterase from human erythrocytes
- Butyrylcholinesterase from human serum
- Acetylcholine chloride
- Butyrylcholine chloride
- BW284c51 (AChE-specific inhibitor)
- Magnesium chloride
- Calcium chloride
- Phosphate buffer saline (0.1 M, pH 7.4)
- Finger-stick blood samples
Procedure:
Preparation of CS/MWCNT/PAnNF Nanocomposite:
- Synthesize polyaniline nanofibers via interfacial polymerization
- Functionalize MWCNTs by acid treatment (3:1 H₂SO₄:HNO₃) for 4 hours
- Prepare chitosan solution (1% w/v) in acetic acid (1% v/v)
- Disperse MWCNTs (2 mg/mL) in chitosan solution by probe sonication
- Mix MWCNT/chitosan suspension with PAnNF dispersion at 1:1 volume ratio
- Stir for 2 hours to form homogeneous nanocomposite
Electrode Modification:
- Clean gold interdigitated electrodes with oxygen plasma treatment
- Drop-cast CS/MWCNT/PAnNF nanocomposite (5 μL) onto electrode surface
- Dry at room temperature for 4 hours followed by 40°C for 30 minutes
- Characterize modified electrode using SEM and electrochemical impedance spectroscopy
Reagent Pad Preparation:
- Prepare outer pretreatment pad by soaking glass fiber pad (Ø 4 mm) in inhibitor solution (BW284c51 for AChE specificity)
- Prepare inner signal generation pad with acetylcholine (for AChE) or butyrylcholine (for BChE) substrate
- Dry pads under vacuum for 2 hours and store with desiccant
Pesticide Detection Assay:
- Apply whole blood sample (10 μL) to outer pretreatment pad
- Assemble sensor with reagent pads in contact with modified electrode
- Connect electrode to Bluetooth resistance meter paired with smartphone
- Measure conductance change over 5-minute interval
- Calculate cholinesterase activity from conductance slope
- Determine pesticide concentration from enzyme inhibition percentage
Data Analysis:
- Transmit resistance measurements to smartphone via Bluetooth
- Calculate cholinesterase activity using calibration curve
- Determine pesticide exposure level based on enzyme inhibition
- Store results with timestamp and geolocation data
Signaling Pathways and Mechanisms
The following diagrams illustrate the signaling pathways and mechanisms for optical and electrochemical biosensors used in pesticide detection.
Diagram 1: Optical transduction signaling pathway for pesticide detection based on enzyme inhibition and nanozyme-catalyzed color development.
Diagram 2: Electrochemical transduction signaling pathway for pesticide detection based on enzyme inhibition and proton-mediated conductance changes.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Essential Research Reagents and Materials for Biosensor Development
| Category | Item | Function | Example Applications |
|---|---|---|---|
| Biological Elements | Acetylcholinesterase | Primary recognition element for OPs | Enzyme inhibition assays [23] [20] |
| Aptamers | Nucleic acid-based recognition elements | Target-specific molecular recognition [19] | |
| Antibodies | Immunoaffinity recognition | Molecular imprinting and immunoassays [25] | |
| Nanomaterials | Graphene Oxide | Fluorescence quenching in FRET assays | Optical aptasensors [19] |
| HOF Nanozymes | Peroxidase-mimicking activity | Colorimetric detection [20] | |
| MWCNT/PAnNF | Conductance-based sensing | Electrochemical biosensors [23] | |
| Gold Nanoparticles | Signal amplification, SERS substrates | Enhanced detection sensitivity [26] | |
| Signal Probes | TMB | Chromogenic substrate | Colorimetric detection [20] |
| Acetylthiocholine | Enzyme substrate | Electrochemical and optical assays [23] [20] | |
| Methylene Blue | Electrochemical redox probe | Voltammetric sensing [26] | |
| Support Materials | Sodium Alginate | Hydrogel matrix formation | Biosensor immobilization [20] |
| Chitosan | Biocompatible polymer matrix | Nanocomposite formation [23] | |
| Gold Interdigitated Electrodes | Transduction platform | Resistive/conductive measurements [23] |
Optical and electrochemical transduction mechanisms offer complementary advantages for smartphone-integrated biosensors targeting pesticide detection. Optical methods, particularly colorimetric approaches using nanozymes, provide visual readouts ideally suited for smartphone camera detection with sensitivity meeting practical requirements. Electrochemical techniques offer inherent advantages for miniaturization and direct electronic integration with smartphone platforms. The choice between these mechanisms depends on specific application requirements including sensitivity needs, sample matrix, instrumentation constraints, and intended user operation. Future developments will likely focus on hybrid approaches combining the visual simplicity of optical detection with the electronic interface capabilities of electrochemical systems, further enhanced by artificial intelligence for signal processing and result interpretation.
The field of biosensing is undergoing a transformative shift, driven by the convergence of artificial intelligence (AI), the Internet of Things (IoT), and cloud connectivity. This synergy is particularly impactful in the development of smartphone-integrated biosensors, creating powerful, decentralized diagnostic and monitoring platforms [27] [7]. These systems are moving analytical capabilities from centralized laboratories directly to the point-of-need, enabling rapid, on-site detection of analytes like pesticides [23] [28] [10]. For researchers focused on visual pesticide detection, this integration addresses critical challenges in sensitivity, specificity, and data management, while opening new avenues for real-time environmental and health monitoring. This document outlines the key technological trends, provides structured experimental data, and details protocols for developing and validating these advanced biosensing systems.
The Converging Technological Landscape
The modern biosensor is no longer a simple transducer but a sophisticated system that leverages advances in multiple domains. The core of this evolution lies in the seamless integration of sensing, computation, and connectivity.
AI-Enhanced Biosensing
Artificial intelligence, particularly machine learning (ML) and deep learning, dramatically improves the analytical performance of optical biosensors. AI algorithms are adept at processing complex, multivariate signal data to enhance sensitivity and specificity [27].
- Intelligent Signal Processing: AI can differentiate target signals from background noise and correct for environmental interferences or matrix effects in complex samples like food [27] [28].
- Multiplexing and Pattern Recognition: For sensors detecting multiple pesticides, AI models can deconvolute overlapping optical signals (e.g., fluorescence, colorimetric) from array-based sensors, enabling simultaneous identification of several analytes [27].
- Predictive Analytics: ML models can predict calibration drift or sensor degradation, prompting recalibration and ensuring data reliability over time [27].
IoT and Connectivity Frameworks
The IoT ecosystem provides the infrastructure for biosensors to become interconnected nodes in a larger network. Billions of connected IoT devices form a foundation for widespread biosensor deployment, with key connectivity technologies including Wi-Fi (32%), Bluetooth (24%), and Cellular IoT (22%) [29].
Smartphone-based biosensors fit into system architectures defined by the location of the biosensing function and data processing [7]:
- Architecture A: On-phone biosensing with local data processing.
- Architecture B: On-phone biosensing with server/cloud processing.
- Architecture C: Off-phone biosensing with local processing (on a dedicated device or smartphone).
- Architecture D: Off-phone biosensing with server processing.
The choice of architecture involves trade-offs between portability, sensing capability, processing power, and data storage [7]. The integration of IoT enables features like real-time data tracking, sharing of results with healthcare providers or regulatory bodies, and large-scale environmental biomonitoring [23].
Edge Computing and Cloud Synergy
A key trend is the move towards edge computing, where data is processed on the device itself or a local gateway rather than being sent entirely to the cloud [30]. For biosensors, this means:
- Ultra-Low Latency: Critical for real-time decision-making, such as immediate alerts for toxic pesticide levels [30].
- Bandwidth and Cost Savings: Only processed results or relevant data subsets are transmitted to the cloud, reducing data transmission loads [30].
- Enhanced Data Privacy: Sensitive information can be analyzed locally, minimizing external data transmission [30].
The cloud complements the edge by providing vast storage for historical data, powerful resources for training complex AI models, and a platform for aggregating data from multiple sensors for large-scale analytics [27] [7].
Quantitative Data in Modern Biosensing
The performance of emerging biosensing platforms is quantified through key analytical parameters. The tables below summarize data from recent implementations relevant to pesticide detection and associated technologies.
Table 1: Analytical Performance of Selected Smartphone-Integrated Biosensors for Pesticide and Contaminant Detection
| Target Analyte | Sensing Platform | Detection Mechanism | Linear Range | Limit of Detection (LOD) | Test Duration | Citation |
|---|---|---|---|---|---|---|
| Organophosphate Pesticides (e.g., Paraoxon-Methyl) | Smartphone/Resistive Nanosensor | AChE inhibition; Conductance change of PAnNF/CNT film | 1 ppt – 100 ppb | 0.304 ppt | ~10 minutes | [10] |
| Acetylcholinesterase (AChE) Activity (OP Exposure Biomarker) | Smartphone/Resistive Nanosensor | Substrate hydrolysis; Conductance change | 2.0–18.0 U/mL | 0.11 U/mL | ~10 minutes | [23] |
| Butyrylcholinesterase (BChE) Activity (OP Exposure Biomarker) | Smartphone/Resistive Nanosensor | Substrate hydrolysis; Conductance change | 0.5–5.0 U/mL | 0.093 U/mL | ~10 minutes | [23] |
| Various Pesticides & Antibiotics | Smartphone/Fluorescent Probe (UOFs) | Ratiometric Fluorescence | N/S (Well below regulatory thresholds) | N/S | ~10 seconds | [28] |
Table 2: IoT Connectivity Landscape Relevant for Distributed Biosensor Networks (2025 Data) [29]
| Connectivity Technology | Share of Global IoT Connections | Key Characteristics & Relevance to Biosensing |
|---|---|---|
| Wi-Fi | 32% | High bandwidth; suitable for fixed or powered sensors in homes, clinics, or labs. |
| Bluetooth | 24% | Low power; ideal for short-range communication between a biosensor and a smartphone. |
| Cellular IoT (5G, LTE-M, NB-IoT) | 22% | Wide area coverage; enables remote biosensing in agricultural or environmental fields. |
| Other (LPWAN, etc.) | 22% | Very low power, long range; for sensors in remote locations with infrequent data transmission. |
Experimental Protocols
This section provides detailed methodologies for implementing and validating key aspects of AI- and IoT-enhanced biosensors for pesticide detection.
Protocol: Development of a Smartphone-Based Resistive Nanosensor for Organophosphate Pesticides
Application: On-site rapid detection of organophosphate pesticides in food and water samples [10].
Principle: The sensor leverages the inhibition of acetylcholinesterase (AChE). In the absence of pesticide, AChE hydrolyzes acetylcholine, releasing protons that dope polyaniline nanofibers (PAnNFs) and increase film conductance. OP pesticides inhibit AChE, reducing the rate of proton generation and the resultant conductance change, which is quantitatively measured [10].
Materials:
- Gold interdigitated electrodes (AuIDEs)
- Chitosan (CS), Multi-walled carbon nanotubes (MWCNTs), Polyaniline nanofibers (PAnNFs)
- Acetylcholinesterase (AChE)
- Acetylcholine (ACh) substrate
- Phosphate buffer saline (PBS, 0.1 M, pH 7.4)
- Smartphone with custom-developed application
- Bluetooth-enabled portable resistance meter
Procedure:
- Nanosensor Fabrication:
- Clean AuIDEs with oxygen plasma.
- Prepare a composite suspension of CS, MWCNTs, PAnNFs, and AChE in acetic buffer.
- Drop-cast the suspension onto the active area of the AuIDE and allow it to dry.
- Integrate reagent pads: an outer glass fiber pad pre-loaded with anti-interference reagents and an inner pad pre-loaded with the ACh substrate [23] [10].
Measurement Setup:
- Connect the fabricated nanosensor to the portable resistance meter.
- Pair the resistance meter with the smartphone app via Bluetooth.
- For calibration, apply 20 μL of standard solutions (or sample extracts) with known pesticide concentrations to the sensor.
- The app initiates resistance measurement and records the data in real-time.
Data Acquisition and Analysis:
- The smartphone app records the resistance change over a fixed period (e.g., 5-10 minutes).
- The rate of resistance change is inversely proportional to AChE activity and directly proportional to pesticide concentration.
- A calibration curve is established within the app by measuring standard solutions.
- Unknown sample concentrations are calculated by the app by interpolating the measured signal against the calibration curve.
Validation:
- Validate the sensor's performance against a standard method like liquid chromatography-mass spectrometry (LC-MS) using spiked food and water samples [10].
- Calculate recovery rates (target: ~98%) and reproducibility (RSD <5%).
Protocol: Implementing AI for Signal Processing in Optical Biosensors
Application: Enhancing the sensitivity and specificity of smartphone-based colorimetric or fluorescent pesticide sensors [27] [28].
Principle: AI models, particularly convolutional neural networks (CNNs), can be trained to analyze images captured by the smartphone camera. They learn to correlate specific visual patterns (hue, intensity, texture) with analyte concentration, compensating for variable lighting conditions and sample impurities.
Materials:
- Smartphone with integrated camera
- A uniform lighting enclosure (to minimize external light variation)
- Trained AI/ML model (e.g., TensorFlow Lite model)
- Sample holder compatible with the smartphone attachment
Procedure:
- Data Collection for Model Training:
- Capture images of the sensor output (e.g., colorimetric test strip, fluorescent probe) across a wide range of known analyte concentrations.
- Vary lighting conditions and use different smartphone models to build a robust dataset.
- Annotate each image with the true concentration.
Model Training and Deployment:
- Pre-process images (e.g., crop region of interest, color space conversion).
- Train a CNN model for regression (to predict concentration) or classification (e.g., safe/unsafe).
- Convert the trained model to a mobile-friendly format (e.g., TFLite) and integrate it into the smartphone application.
In-Field Analysis:
- The user places the reacted sensor in the designated holder.
- The smartphone app automatically captures an image.
- The integrated AI model processes the image and outputs the predicted concentration or classification in real-time.
- Results, along with timestamp and geolocation, can be stored locally or uploaded to a cloud database via IoT connectivity.
Protocol: Deployment and Data Management via IoT/Cloud Architecture
Application: Enabling large-scale, distributed monitoring and real-time data tracking for pesticide exposure or environmental contamination [23] [7].
Principle: Biosensor data is transmitted from the smartphone to a cloud platform, where it is aggregated, stored, and made accessible for visualization and further analysis, facilitating remote monitoring and population-level studies.
Materials:
- Smartphone-integrated biosensor
- Cloud computing service (e.g., Google Cloud Platform, AWS)
- Database (e.g., SQL, NoSQL)
- Web interface for data visualization
Procedure:
- System Architecture Setup (Architecture D [7]):
- Configure the smartphone app to transmit results (measurement, timestamp, device ID, location) to a cloud database via a secure API upon test completion. Connectivity can be Wi-Fi or cellular [29].
- Set up the cloud database to receive and store data from multiple users/devices.
Data Processing and Storage:
- Implement cloud functions to validate incoming data and trigger alerts if measurements exceed predefined thresholds (e.g., pesticide regulatory limits).
- Store raw and processed data in a structured format for long-term access.
Visualization and Sharing:
- Develop a web-based dashboard that displays aggregated data, trends over time, and geographic distribution of results.
- Implement secure, role-based access to the dashboard for researchers, public health officials, or individual users.
- The system can be designed to automatically generate and share reports with relevant stakeholders [23].
Signaling Pathways and Workflows
The following diagrams illustrate the logical workflow of an integrated biosensing system and the molecular signaling principle of a common pesticide detection method.
Diagram 1: Integrated AIoT Biosensing Workflow
Diagram 2: AChE Inhibition Biosensing Principle
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Smartphone-Based Resistive Biosensor Development
| Item / Reagent | Function / Role in the Experiment | Exemplary Specifications / Notes |
|---|---|---|
| Acetylcholinesterase (AChE) | Biological recognition element; catalyzes substrate hydrolysis. | Source: Electric eel or recombinant. Activity >1000 U/mg. Stability under storage conditions is critical. |
| Polyaniline Nanofibers (PAnNFs) | Transducer material; conductance is modulated by proton doping from the enzymatic reaction. | High surface-to-volume ratio enhances sensitivity. Synthesized via oxidative polymerization [23] [10]. |
| Carbon Nanotubes (CNTs) | Nanomaterial enhancing electron transfer and providing a high-surface-area matrix for enzyme immobilization. | Multi-walled (MWCNTs) or single-walled (SWCNTs). Functionalized (e.g., carboxylated) for better dispersion and biocompatibility [23] [10]. |
| Gold Interdigitated Electrode (AuIDE) | Platform for the nanosensor film; interdigitated structure maximizes contact area for sensitive resistance measurement. | Standard finger width/spacing of 10 μm. Requires cleaning (e.g., oxygen plasma) before modification. |
| Chitosan (CS) | Biopolymer used for enzyme immobilization; provides a biocompatible, porous matrix. | High degree of deacetylation. Forms a stable hydrogel in mild acidic conditions for encapsulating AChE/CNT/PAnNF [23] [10]. |
| Acetylcholine (ACh) / Acetylthiocholine (ATCh) | Enzyme substrate; hydrolysis produces protons (ACh) or thiocholine (ATCh), leading to the measurable signal. | ACh for resistive sensors; ATCh for electrochemical sensors. Stability and purity are important for reproducible kinetics. |
| Smartphone & Mobile App | Serves as the user interface, data processor, and communication hub. | App developed in Android/iOS to control measurement, run AI models, display results, and manage IoT data transmission [23] [7]. |
| Portable Resistance Meter | Measures the conductance change of the nanosensor film. | Bluetooth-enabled for wireless communication with the smartphone. Requires stable baseline and high-resolution measurement. |
Implementing Visual Detection: From Probe Design to Real-World Application
The detection of pesticide residues in food and environmental samples is a critical global challenge, necessitating the development of rapid, sensitive, and field-deployable analytical technologies. Traditional methods, such as gas chromatography and high-performance liquid chromatography, offer high precision but are hampered by their high cost, operational complexity, and lack of portability for on-site analysis [1]. In response, smartphone-integrated biosensors have emerged as a transformative platform for point-of-need testing, combining the powerful processing, imaging, and connectivity of consumer devices with advanced biochemical sensing principles [31] [32].
Among the most promising advancements in this field are probe technologies based on uranium-organic frameworks (UOFs) and ratiometric fluorescence. UOFs are a class of metal-organic frameworks that leverage uranyl ions as the metal center, offering exceptional water stability, strong luminescence, and unique photocatalytic properties [28] [33]. Concurrently, ratiometric fluorescence sensing employs the ratio of fluorescence intensities at two different wavelengths, providing a built-in calibration that minimizes environmental interference and significantly enhances measurement accuracy and sensitivity compared to single-intensity probes [31] [34]. The synergy of these technologies with smartphone-based detection creates a powerful tool for the visual, quantitative, and on-site screening of hazardous pesticides, paving the way for a new generation of food safety monitoring systems [28] [35].
Uranium-Organic Frameworks (UOFs) as Sensing Probes
Uranium-organic frameworks are a specific type of MOF characterized by their unique photophysical and structural properties. The uranyl ions (UO₂²⁺) impart several key advantages for sensing applications:
- Enhanced Rigidity and Luminescence: The incorporation of uranium centers and a heterometallic design in UOFs results in enhanced structural rigidity, defined pore structures, and strong luminescence, which are crucial for sensitive detection [28].
- Photoactive Properties: Uranyl ions possess a high oxidation potential (
E₀ = 2.6 V) and can participate in ligand-to-metal charge transfer (LMCT) processes. This allows UOFs to act not only as sensors but also as photocatalysts for the degradation of pollutants, a property explored in environmental remediation [33]. - Tailorable Specificity: Researchers can synthesize UOFs with specific structures and luminescent properties by selecting appropriate organic linkers and synthesis conditions. For instance, three distinct heterometallic UOFs were tailored to respond to different pesticides and antibiotics through unique fluorescence responses, enabling selective detection [28].
Principles of Ratiometric Fluorescence Sensing
Ratiometric fluorescence is a self-referencing technique that significantly improves the reliability of fluorescence-based assays. Its core principle and advantages are:
- Self-Calibration Mechanism: This method uses the change in fluorescence intensity at two different emission wavelengths. The ratio of these two intensities (
I₁ / I₂) is used as the analytical signal, which automatically corrects for variations in experimental conditions such as probe concentration, excitation light intensity, and environmental noise [28] [31]. - Superior Sensitivity and Accuracy: Probes designed with two emissions that change in opposite directions (one increasing and the other decreasing) in response to an analyte provide the highest sensitivity. This opposite response amplifies the change in the ratiometric signal, leading to lower limits of detection [31].
- Visual and Qualitative Analysis: The distinct color changes under UV light that often accompany ratiometric sensing allow for qualitative analysis by the naked eye, which can then be quantified precisely using a smartphone's camera and a dedicated application [36] [34].
Experimental Protocols
Protocol 1: Synthesis of Heterometallic UOFs for Pesticide Detection
This protocol is adapted from the work of Yang et al., which focused on developing a smartphone-integrated sensor for pesticides and antibiotics [28].
- Objective: To synthesize heterometallic uranium-organic frameworks (UOFs) with strong luminescence and tailored specificity for use as fluorescent probes.
- Materials:
- Metal Precursors: Uranyl salt (e.g.,
UO₂(NO₃)₂·6H₂O) and salts of secondary metals (e.g.,Ca²⁺,Sr²⁺,Ba²⁺). - Organic Linkers: Tricarboxylic acid ligands (e.g., imidazole-based ligands).
- Solvent:
N,N'-Dimethylformamide (DMF), deionized water.
- Metal Precursors: Uranyl salt (e.g.,
- Procedure:
- Dissolution: Dissolve the uranyl salt, the secondary metal salt, and the organic linker in a mixture of DMF and water in a defined molar ratio.
- Solvothermal Reaction: Transfer the solution to a Teflon-lined autoclave. Heat the autoclave to 120-150 °C and maintain this temperature for 24-48 hours to allow for slow crystal growth.
- Product Recovery: After the reaction vessel has cooled to room temperature, collect the resulting crystalline product via centrifugation.
- Purification: Wash the crystals repeatedly with fresh DMF and ethanol to remove unreacted precursors and impurities.
- Activation: Finally, dry the purified UOF crystals under vacuum at an elevated temperature (e.g., 100 °C) for 12 hours to remove solvent molecules from the pores, activating them for sensing applications.
- Key Notes: The specific structure and luminescent properties of the UOF are highly dependent on the reaction temperature, time, and the metal-to-ligand ratio. These parameters must be optimized for the target analytes.
Protocol 2: Smartphone-Based Ratiometric Fluorescence Detection of Pesticides
This protocol details the use of a synthesized UOF probe for the actual detection of pesticides, integrated with a smartphone for readout [28] [34].
- Objective: To quantitatively detect pesticide residues in food samples using a UOF-based ratiometric fluorescent probe and a smartphone-based fluorospectrophotometer.
- Materials:
- Probe: Synthesized UOF particles (from Protocol 1), suspended in a suitable buffer.
- Samples: Food extracts (e.g., from vegetables, fruits).
- Equipment: Custom smartphone fluorospectrophotometer (SBS), UV light source (365 nm), cuvette.
- Software: Custom Android application (e.g., SBS-App) for spectral analysis [34].
- Procedure:
- Sample Preparation: Homogenize the food sample (e.g., 1.0 g of apple or cabbage) and extract pesticides using a solvent like acetonitrile. Filter the extract to remove particulate matter.
- Detection Reaction: In a cuvette, mix 100 µL of the filtered food extract with 900 µL of the UOF probe suspension. Allow the mixture to react for 10 seconds.
- Signal Acquisition: Place the cuvette in the holder of the smartphone-based fluorospectrophotometer. Irradiate the mixture with a 365 nm UV LED. Use the smartphone's camera and the SBS-App to capture the fluorescence emission spectrum across the visible range (380-760 nm).
- Data Processing: The SBS-App automatically converts the captured image into a fluorescence spectrum and calculates the intensity ratio (
I₁ / I₂) at two predetermined emission wavelengths. - Quantification: The concentration of the target pesticide in the sample is determined by interpolating the measured intensity ratio against a pre-established calibration curve.
- Key Notes: The entire process, from sample reaction to quantitative result, can be completed in under 30 seconds. The smartphone app is critical for converting raw images into spectral data and performing the ratiometric calculation, which minimizes user-induced errors [28] [34].
Signaling Pathways and Workflows
The detection mechanism for pesticides, particularly organophosphorus pesticides (OPs), can be based on enzyme inhibition. The following diagram illustrates the signaling pathway for this type of sensor.
The experimental workflow, from probe preparation to final analysis, is a multi-step process that integrates chemistry, materials science, and smartphone technology, as outlined below.
Research Reagent Solutions and Materials
The following table details key reagents and materials essential for developing and implementing UOF-based ratiometric fluorescence sensors.
Table 1: Essential Research Reagents and Materials for UOF-based Ratiometric Sensing
| Item Name | Function/Description | Key Characteristics |
|---|---|---|
Uranyl Salts (e.g., UO₂(NO₃)₂·6H₂O) |
Metal precursor for UOF synthesis | Provides the photoactive UO₂²⁺ center; defines framework topology and luminescence [28] [33]. |
| Tricarboxylic Acid Ligands | Organic linker for UOF synthesis | Connects metal nodes to form porous frameworks; functional groups influence specificity and stability [28] [33]. |
Heterometallic Salts (e.g., CaCl₂, Sr(NO₃)₂) |
Co-metal precursor for UOF synthesis | Enhances structural rigidity and tailors the luminescent response to specific analytes [28]. |
| Smartphone Fluorospectrophotometer (SBS) | Portable detection device | Custom attachment with UV LED, diffraction grating, and cuvette holder; uses smartphone CMOS for spectral capture [34]. |
| SBS-App | Data processing software | Custom Android/iOS application for converting camera images to spectra and calculating ratiometric values [34]. |
| Acetylcholinesterase (AChE) | Biological recognition element | Enzyme whose inhibition by OPs is the basis for the sensing mechanism in many ratiometric assays [36] [34]. |
Performance Data and Comparison
The performance of UOF and other ratiometric probes for pesticide detection is quantified by parameters such as detection limit, linear range, and recovery rate in real samples. The data below summarizes the capabilities of these advanced sensors.
Table 2: Performance Comparison of Ratiometric Fluorescence Sensors for Pesticide Detection
| Detection Platform / Probe | Target Pesticide | Linear Range | Detection Limit | Application in Real Samples |
|---|---|---|---|---|
| Heterometallic UOFs [28] | Multiple antibiotics & pesticides | Not specified | Below regulatory thresholds | Vegetables, animal products |
| FRET-based Aptasensor (MWCNTs/AuNPs) [37] | Acetamiprid (ACE) | 4 – 40 pM | 2.8 pM | Bell pepper |
| MnO₂ Nanosheet Sensor (SC & AR) [36] | Organophosphorus (e.g., DDVP) | 5.0 pg/mL – 500 ng/mL | 1.6 pg/mL | Apple, cabbage |
| Smartphone Fluorospectrophotometer (CDs & QDs) [34] | Chlorpyrifos | 0.5 – 50 ng/mL | 0.42 ng/mL | Apple, cabbage |
The high sensitivity and selectivity of these sensors are further validated through recovery studies in complex food matrices. For instance, the smartphone fluorospectrophotometer (SBS) achieved recovery rates of 94.6% to 105.8% for chlorpyrifos in apple and cabbage samples, demonstrating accuracy comparable to the standard GC-MS method [34]. Similarly, the FRET-based aptasensor for acetamiprid showed efficient performance in bell pepper samples, confirming its applicability in agricultural products [37].
Colorimetric and Fluorometric Assay Development for Pesticide Sensing
The increasing use of organophosphorus (OP) and carbamate (CM) pesticides in modern agriculture poses significant threats to human health and environmental safety. These compounds function by inhibiting acetylcholinesterase (AChE), an enzyme crucial for proper nervous system function, leading to potential neurological dysfunction and other health issues upon chronic exposure [38] [39]. While conventional methods like gas chromatography and high-performance liquid chromatography offer high sensitivity, they require sophisticated instrumentation, extensive sample preparation, and lack suitability for rapid on-site screening [1] [39]. Consequently, developing simple, rapid, and reliable detection methods has become imperative for environmental monitoring and food safety control.
Biosensing technologies, particularly colorimetric and fluorometric assays, have emerged as viable alternatives to conventional methods, offering exceptional sensitivity, rapid response, and ease of operation [1]. The integration of these assays with smartphones further enhances their potential for point-of-care testing, enabling real-time, on-site detection of pesticide residues [20] [28]. This application note details the development of robust colorimetric and fluorometric assays for pesticide sensing, framed within a broader research context focused on smartphone-integrated biosensors for visual pesticide detection.
Key Sensing Mechanisms and Performance Comparison
Contemporary research has explored various sensing mechanisms for pesticide detection, primarily based on enzyme inhibition principles or nanozyme-enhanced catalysis. The table below summarizes the key characteristics and analytical performance of different assay types.
Table 1: Comparison of Colorimetric and Fluorometric Assays for Pesticide Detection
| Assay Type | Sensing Mechanism | Target Pesticide | Limit of Detection (LOD) | Analysis Time | Key Features |
|---|---|---|---|---|---|
| Colorimetric (Nanozyme-enhanced) [38] | Enhancement of oxidase-mimicking activity of cube-shape Ag₂O | Dimethoate (Organophosphorus) | 14 μg·L⁻¹ | < 10 minutes | Simple, rapid, reliable; Does not require H₂O₂ |
| Colorimetric (Enzyme Inhibition) [39] | Inhibition of cricket cholinesterase | Organophosphates & Carbamates | 0.002–0.877 ppm | Optimized at 5 min | Low-cost, uses widely available cricket enzyme |
| Fluorometric/Colorimetric Bimodal [40] | Enzyme-triggered decomposition of AuNCs-MnO₂ nanocomposite | Carbaryl (Carbamate) | 0.125 μg·L⁻¹ | - | Dual-output for self-verification; High sensitivity and anti-interference |
| Smartphone-assisted Hydrogel Biosensor [20] | HOF nanozyme-based inhibition assay | Chlorpyrifos (Organophosphorus) | 3.04 ng/mL | - | Portable, on-site detection; Robust stability |
Detailed Experimental Protocols
Protocol 1: Colorimetric Detection of Dimethoate Using Cube-Shape Ag₂O Nanozyme
This protocol outlines a strategy based on enhancing the oxidase-mimicking activity of cube-shape Ag₂O for rapid dimethoate detection [38].
Materials and Reagents
- AgNO₃ powder
- NaOH
- 3,3′,5,5′-Tetramethylbenzidine (TMB)
- Dimethoate standard
- Acetate buffer (0.2 M, pH 4.0)
- Absolute ethanol
Step-by-Step Procedure
- Synthesis of Cube-Shape Ag₂O Particles: Prepare cube-shape Ag₂O particles according to the previously reported method. Briefly, mix solutions of AgNO₃ and NaOH, then stir vigorously at room temperature. Collect the resulting cube-shape Ag₂O particles by centrifugation, wash with ethanol and water, and dry [38].
- Preparation of Detection System: In a standard cuvette or microplate well, mix the following:
- Acetate buffer (0.2 M, pH 4.0): 500 μL
- TMB solution (2.0 mM): 100 μL
- Cube-shape Ag₂O suspension (50 μg·mL⁻¹): 100 μL
- Varying concentrations of dimethoate standard or sample extract.
- Catalytic Reaction and Measurement: Incubate the reaction mixture at 35 °C for 10 minutes. The presence of dimethoate enhances the oxidase-mimicking activity of Ag₂O, catalyzing the oxidation of TMB to a blue product (oxTMB).
- Signal Acquisition: Measure the absorbance of the solution at 652 nm using a spectrophotometer or a smartphone-based colorimetric detection system. The increase in absorbance is directly proportional to the dimethoate concentration.
Signaling Pathway Workflow
Protocol 2: Fluorometric/Colorimetric Bimodal Detection of Carbaryl Using AuNCs-MnO₂ Nanocomposite
This protocol describes a dual-output sensing platform for carbaryl based on an enzyme-triggered decomposition of a gold nanoclusters-anchored MnO₂ nanocomposite [40].
Materials and Reagents
- Acetylcholinesterase (AChE)
- Choline Oxidase (ChOx)
- Acetylcholine (ACh) iodide
- Bovine Serum Albumin (BSA)
- HAuCl₄·xH₂O
- MnCl₂
- Carbaryl standard
Step-by-Step Procedure
- Synthesis of AuNCs-MnO₂ Nanocomposite: Prepare BSA-stabilized gold nanoclusters (AuNCs) via a biomimetic synthesis. Then, synthesize the AuNCs-MnO₂ composite using a co-template hydrothermal reaction by mixing the as-prepared AuNCs with MnCl₂ and incubating at elevated temperature [40].
- Establishing the Baseline Signal: In a quartz cuvette, mix:
- Phosphate buffer (e.g., 20 mM, pH 7.4)
- AuNCs-MnO₂ nanocomposite
- AChE and ChOx enzymes
- Substrate ACh The H₂O₂ generated from the enzymatic cascade decomposes the MnO₂, causing the recovery of AuNCs' fluorescence and a change in the solution color.
- Inhibition Assay for Carbaryl Detection: Pre-incubate the AChE enzyme with different concentrations of carbaryl standard or sample for a set time (e.g., 10-15 minutes). Then, add this mixture to the system described in Step 2. Carbaryl inhibits AChE, reducing H₂O₂ production, thus suppressing the decomposition of MnO₂. This results in continued fluorescence quenching and a diminished color change.
- Dual-Mode Signal Acquisition:
- Fluorometric: Measure the fluorescence recovery at the emission wavelength of AuNCs (e.g., ~650 nm with excitation at ~370 nm).
- Colorimetric: Measure the absorbance change of the MnO₂ nanocomposite or the colored enzymatic product.
Bimodal Sensing Logic
The Scientist's Toolkit: Essential Research Reagents
The development and execution of these advanced biosensors rely on several key classes of materials and reagents. The following table details these essential components and their functions.
Table 2: Key Research Reagent Solutions for Pesticide Biosensor Development
| Reagent Category | Specific Examples | Function in the Assay |
|---|---|---|
| Biological Recognition Elements | Acetylcholinesterase (AChE), Butyrylcholinesterase (BChE), Cricket Cholinesterase [41] [39] | High-specificity binding and inhibition by target organophosphate and carbamate pesticides. |
| Nanozymes & Functional Nanomaterials | Cube-shape Ag₂O [38], HOF-based nanozymes (e.g., Hemin@HOF) [20], AuNCs-MnO₂ nanocomposite [40] | Mimic natural enzyme activity; act as signal amplifiers or reporters; enhance catalytic stability and sensitivity. |
| Chromogenic/Fluorogenic Substrates | 3,3',5,5'-Tetramethylbenzidine (TMB) [38] [20], Acetylthiocholine (ATCh) / DTNB [39] | Generate measurable colorimetric or fluorometric signals upon enzymatic or nanozyme-catalyzed reaction. |
| Signal Probes & Labels | Gold Nanoclusters (AuNCs) [40], Uranium-Organic Frameworks (UOFs) [28] | Serve as highly sensitive fluorescent reporters for ratiometric or intensity-based detection. |
| Smartphone Integration Components | Hydrogel matrix [20], Custom mobile application, Cuvette adapter | Enable solid-phase sensing, portability, and quantitative colorimetric/fluorometric analysis for on-site testing. |
The detailed protocols for colorimetric and fluorometric assays presented herein provide robust methodologies for sensitive pesticide detection. The integration of novel nanomaterials, such as nanozymes and fluorescent nanocomposites, significantly enhances analytical performance. Furthermore, the compatibility of these assays with smartphone-based readout systems paves the way for developing powerful, portable, and user-friendly tools for on-site pesticide monitoring, contributing substantially to food safety and environmental protection. Future work will focus on expanding multi-analyte detection capabilities and further optimizing sensor integration with mobile technology.
The reliable detection of pesticide residues in complex environmental and agricultural matrices is a critical challenge for ensuring food safety and environmental health. Traditional laboratory methods, while sensitive, are often ill-suited for rapid, on-site screening due to their cost, operational complexity, and lack of portability [42]. The emergence of smartphone-integrated biosensors presents a transformative solution, offering the potential for decentralized, visual, and quantitative analysis [28] [43]. These systems leverage the computational power, connectivity, and high-resolution cameras of smartphones, turning them into portable diagnostic tools [43].
A significant hurdle in this field is the sample preparation of complex matrices—food, water, and soil. These samples contain interferents that can severely affect the sensitivity and accuracy of biosensors. This application note provides detailed protocols for the preparation and analysis of these matrices, specifically tailored for smartphone-based visual biosensing platforms. The focus is on practical, field-deployable methodologies that enable researchers to transition from laboratory-based assays to real-world application.
Sample Preparation Protocols
Proper sample preparation is fundamental for minimizing matrix effects and achieving reliable detection with biosensors. The following protocols are designed for compatibility with subsequent smartphone-based analysis.
Food Sample Preparation (Fruits and Vegetables)
The primary goal for food samples is the extraction of pesticide residues from the heterogeneous food surface and pulp, while removing interfering compounds like pigments and organic acids.
Materials:
- Homogenizer (e.g., blender or stomacher)
- Centrifuge and centrifuge tubes
- Filtration units (0.45 μm syringe filters)
- Extraction solvent: Acetone, ethyl acetate, or acetonitrile
- Buffers: Phosphate Buffered Saline (PBS, pH 7.4)
Procedure:
- Representative Sampling: Select and thoroughly wash the fruit or vegetable with tap water to remove soil and debris. Air-dry at room temperature.
- Comminution: Precisely weigh 10.0 g of the edible portion and homogenize it into a puree using a blender.
- Solvent Extraction: Transfer the puree into a 50 mL centrifuge tube. Add 20 mL of extraction solvent (e.g., acetonitrile). Vortex vigorously for 2 minutes to ensure complete solvent-sample interaction.
- Centrifugation: Centrifuge the mixture at 8,000 rpm for 10 minutes to separate the solid plant material from the liquid extract containing the pesticides.
- Filtration: Carefully collect the supernatant and pass it through a 0.45 μm filter to remove any remaining particulate matter.
- Dilution and pH Adjustment: Dilute a known volume of the filtrate with PBS to a ratio of 1:10. This step is crucial for reducing the solvent strength and adjusting the pH to be compatible with the biochemical reagents (e.g., acetylcholinesterase) used in the biosensor [42].
Water Sample Preparation
Surface and groundwater samples may contain suspended solids and dissolved organic matter that can interfere with detection.
Materials:
- Filtration apparatus and 0.22 μm membrane filters
- pH meter and adjustment solutions (e.g., HCl, NaOH)
Procedure:
- Collection: Collect water samples in clean glass containers. For groundwater, allow the pump to run for several minutes before sampling to ensure a representative sample.
- Filtration: Filter the water sample through a 0.22 μm membrane filter to remove suspended particles and microorganisms.
- pH Adjustment: Measure the pH of the filtered sample. If necessary, adjust to pH 7.4 using dilute HCl or NaOH to ensure optimal enzyme activity in the biosensing system [42].
- Analysis: The filtered, pH-adjusted sample is now ready for direct analysis without further dilution, unless the pesticide concentration is expected to be very high.
Soil Sample Preparation
Soil is a highly complex matrix; the protocol aims to extract pesticides adsorbed to soil particles while co-extracting the least amount of humic substances.
Materials:
- Soil probe or auger
- Siever (2-mm mesh)
- Mechanical shaker
- Solid-phase extraction (SPE) cartridges (e.g., C18) for clean-up
Procedure:
- Composite Sampling: Using a soil probe, collect 15-20 sub-samples from a defined area (e.g., a hectare) at a consistent depth (e.g., 0-15 cm for vegetable crops) [44]. Combine and mix these sub-samples thoroughly to create a representative composite sample.
- Preparation: Air-dry the composite sample at room temperature. Crush any large clumps and sieve it through a 2-mm mesh to remove stones and roots [44] [45].
- Solvent Extraction: Precisely weigh 5.0 g of the sieved soil into a 50 mL centrifuge tube. Add 20 mL of an appropriate solvent (e.g., acetone-hexane mixture). Place the tube on a mechanical shaker and agitate for 30 minutes.
- Centrifugation and Collection: Centrifuge the mixture at 5,000 rpm for 5 minutes. Carefully collect the supernatant.
- Clean-up (if required): For soils with high organic content, pass the extract through a C18 SPE cartridge to remove co-extracted humic acids, which can foul the sensor.
- Reconstitution: Evaporate the clean extract to dryness under a gentle stream of nitrogen. Reconstitute the residue in 1 mL of PBS buffer for analysis [46].
Analysis via Smartphone-Integrated Biosensors
The prepared samples can be analyzed using various smartphone-integrated biosensor formats. Two prominent approaches are detailed below.
Fluorescent Probe-Based Detection
This method utilizes the high-resolution camera of a smartphone to capture changes in fluorescence intensity.
Materials:
- Smartphone with a fluorescent probe analysis app
- Dark box to exclude ambient light
- Uranium-Organic Frameworks (UOFs) or other fluorescent probes [28]
- Microplate or test strip holder
Procedure:
- Incubation: Mix a fixed volume of the prepared sample extract with the UOF fluorescent probe in a microplate well or on a test strip.
- Reaction: Allow the mixture to incubate. Pesticides like organophosphates can quench (reduce) the fluorescence of the probe. The reaction is very fast, with reported detection times as low as 10 seconds [28].
- Imaging and Analysis: Place the plate or strip inside the dark box. Using the smartphone app, capture an image of the fluorescence emission. The app uses a ratiometric sensing strategy, analyzing the intensity at two emission wavelengths to self-calibrate and minimize environmental noise, providing a highly selective result [28].
Colorimetric Enzyme-Based Detection
This method is based on the inhibition of the enzyme acetylcholinesterase (AChE) by pesticides, which prevents a color-changing reaction.
Materials:
- Smartphone with a colorimetric analysis app
- AChE-immobilized test card (e.g., on electrospun nanofibers) [42]
- Substrate card impregnated with indoxyl acetate (IA)
Procedure:
- Application: Apply a drop (e.g., 50 μL) of the prepared sample onto the Enzyme Card (EC).
- Inhibition: Allow the sample to incubate on the EC for a defined inhibition time (e.g., 10 minutes). If pesticides are present, they will inhibit the AChE enzyme immobilized on the card.
- Color Development: Bring the inhibited EC into contact with the Substrate Card (SC). The active AChE on the card hydrolyzes IA, producing a blue-green color. The intensity of this color is inversely proportional to the pesticide concentration.
- Analysis: Capture an image of the developed card using the smartphone app. The app converts the color intensity into a quantitative value for the pesticide concentration. This entire process can be completed in approximately 11 minutes [42].
Performance Data and Comparison
The following tables summarize the analytical performance of smartphone-based detection methods for pesticides across different matrices, as reported in recent literature.
Table 1: Analytical Performance of Smartphone-Based Biosensors for Pesticide Detection
| Detection Method | Target Pesticides | Sample Matrix | Detection Limit | Total Analysis Time | Reference |
|---|---|---|---|---|---|
| Fluorescent UOF Probe | Multiple antibiotics & pesticides | Food samples | Varies by compound | ~10 seconds | [28] |
| Colorimetric AChE Nanofiber Card | Phoxim, Methomyl | Fruits & Vegetables | 0.007 mg/L, 0.10 mg/L | ~11 minutes | [42] |
| Dual-Phase Visual Emitters | Trifluralin, Fenitrothion | Soil, Fruits, Vegetables | ~180 nM (for Trifluralin) | Rapid (specific time not given) | [46] |
Table 2: Key Reagents and Materials for Smartphone-Integrated Biosensing
| Research Reagent / Material | Function / Explanation | Application in Protocol |
|---|---|---|
| Acetylcholinesterase (AChE) | Enzyme inhibited by organophosphate & carbamate pesticides; core of the recognition element. | Immobilized on test cards for colorimetric detection [42]. |
| Uranium-Organic Frameworks (UOFs) | Fluorescent probes that change emission intensity upon binding with pesticides. | Used as a ratiometric fluorescent sensor for rapid screening [28]. |
| Electrospun Nanofiber Mat (e.g., PVA/CA) | High-surface-area substrate for enzyme immobilization; enhances stability and sensitivity. | Serves as the solid support for the AChE enzyme card [42]. |
| Indoxyl Acetate (IA) | Enzyme substrate; hydrolysis by AChE produces a colored product (blue-green) for visual detection. | Impregnated on the substrate card for color development [42]. |
| Phosphate Buffered Saline (PBS) | Provides a stable pH environment crucial for maintaining biomolecule (enzyme) activity. | Used for diluting and reconstituting sample extracts [42]. |
Workflow and Signaling Pathways
The following diagrams illustrate the logical workflow for sample analysis and the mechanism of action for the primary detection methods.
Diagram 1: Sample Analysis Workflow. This flowchart outlines the sample preparation and analysis pathway for food, water, and soil matrices, culminating in smartphone-based detection.
Diagram 2: Detection Mechanisms. This diagram compares the two primary signaling mechanisms used in smartphone-based pesticide detection: enzyme inhibition for colorimetric assays and fluorescence quenching for probe-based assays.
The integration of smartphones into biosensing platforms represents a paradigm shift in pesticide detection technology. By leveraging global smartphone connectivity, these systems transform sophisticated chemical analysis from a laboratory-bound procedure into a portable, accessible, and cost-effective tool for on-site monitoring [5]. This integration is built upon three core technological pillars: purpose-built hardware attachments that condition physical or optical signals, sophisticated mobile applications that guide users and process data, and advanced image processing algorithms that extract quantitative information from visual responses [6]. This framework enables the detection of pesticide residues with sensitivities approaching those of traditional laboratory instruments like gas chromatography and mass spectrometry, but with unprecedented speed and field-deployment capability [1] [35]. The following application notes detail the protocols and methodologies that underpin this transformative technology.
Detection Modalities and Hardware Configurations
Smartphone-based detection primarily utilizes optical methods, capitalizing on the device's built-in camera and processing power. The choice of detection modality directly influences the design of the hardware attachment and the accompanying software algorithms.
Table 1: Comparison of Smartphone-Based Detection Modalities for Pesticide Analysis
| Detection Modality | Measurement Principle | Typical Hardware Attachment | Key Advantages | Reported Performance |
|---|---|---|---|---|
| Colorimetry [47] [6] | Measures color intensity or changes in RGB values from a reaction. | Simple dark box to minimize ambient light; may include a uniform LED light source. | Low cost, simplicity, rapid results (e.g., 3 min [47]), high user-friendliness. | Detection of Carbosulfan with 96.7% accuracy [48]; quantification of antioxidants at 0.1 μM [47]. |
| Fluorescence Spectroscopy [28] | Measures intensity changes in emitted light from fluorescent probes. | Attachment with a specific excitation LED and an emission filter. | Very high sensitivity (picomolar range), selectivity through specific probes. | Uranium-organic framework (UOF) probes enable detection in 10 seconds [28]. |
| Spectrophotometry [48] | Measures light absorption across wavelengths to identify molecular fingerprints. | 3D-printed attachment with a diffraction grating to generate a spectrum. | Provides richer data than colorimetry, suitable for machine learning analysis. | Accurately predicted KMnO4 concentration with 98.5% accuracy [48]. |
| Molecularly Imprinted Polymer (MIP) Sensors [35] | Uses synthetic polymers as custom-made recognition sites for specific pesticides. | Can be coupled with colorimetric, electrochemical, or optical readouts. | High stability and selectivity in complex sample matrices like tea [1] [35]. | Effectively recognizes specific molecules for rapid analysis [35]. |
Experimental Protocol: Colorimetric Detection with a Smartphone
Principle: This protocol utilizes a catalyst (e.g., PDFeNi foam [47]) with peroxidase-like activity to oxidize a colorless substrate (e.g., TMB) into a colored product in the presence of pesticides. The pesticide concentration is inversely correlated to the color intensity, as pesticides inhibit the catalytic activity.
Materials:
- Smartphone with a high-resolution camera and the custom-built application installed.
- Dark Box: A simple 3D-printed or fabricated enclosure to house the sample and block ambient light.
- Reference Color Card (optional but recommended for illumination correction [6]).
- Catalyst: Polydopamine-decorated FeNi (PDFeNi) foam [47] or other nanozyme.
- Chromogenic Substrate: TMB, OPD, or 4-AT solution.
- Microplate or Test Strips for holding the reaction mixture.
Procedure:
- Sample Preparation: Extract the pesticide from the target sample (e.g., tea leaves, vegetables) using a suitable solvent. The sample may require filtration or dilution.
- Colorimetric Reaction:
- In a well of a microplate or on a test strip, mix the sample extract, the chromogenic substrate (e.g., TMB), and the PDFeNi foam catalyst.
- Incubate the mixture for a fixed time (e.g., 3 minutes [47]) at room temperature.
- Image Acquisition:
- Place the reacted sample inside the dark box attachment aligned with the smartphone camera.
- Position the optional reference color card next to the sample within the camera's field of view.
- Using the smartphone application, capture an image under standardized lighting conditions provided by an integrated LED.
- Image Analysis & Quantification:
- The application automatically selects the region of interest (ROI) using edge detection and color thresholding.
- It corrects for illumination variations using the Gray World algorithm or the reference color card to calculate a 3x3 color conversion matrix [6].
- The average RGB or grayscale values from the ROI are fed into a pre-trained machine learning model (e.g., Support Vector Regression - SVR) to output the pesticide concentration.
The workflow for this protocol is summarized in the following diagram:
Mobile Application Architecture and Workflow
The mobile application serves as the user interface and the computational engine. Its architecture is designed to be cross-platform and user-friendly, requiring minimal technical expertise from the end-user, such as a farmer or field inspector [6].
Front-End Architecture and User Workflow
Most applications, like the one developed by BAID-China, use frameworks like Flutter to ensure native performance on both iOS and Android from a single codebase [6]. The key modules and their functions are:
- Camera Module: Utilizes the device's native camera API. It often includes an overlay to guide the user in aligning the test strip or sample.
- Image Preview and Cropping: Allows for automatic ROI selection via contour detection, with an option for manual adjustment to ensure accuracy.
- User Interface (UI): Follows Material Design principles with large buttons and clear instructions for use in non-laboratory environments (e.g., in a field) [6].
The complete user journey within the application is a streamlined, step-by-step process:
Back-End Algorithms and Data Processing
Once an image is captured, the back-end processing involves several critical steps to ensure accuracy and robustness against environmental variables [6] [48].
- Region of Interest (ROI) Selection and Extraction: The application uses algorithms like edge detection and color threshold processing to isolate the area containing the color reaction from the background.
- Illumination Correction: This is a crucial step to mitigate variations in lighting conditions. Two common strategies are:
- Gray World Algorithm: Assumes the average color of the scene is neutral gray and adjusts the RGB channels proportionally [6].
- Color Card Calibration: If a reference color card is present in the image, a 3x3 color conversion matrix is calculated to map the captured colors to their reference values, providing a more robust correction [6].
- Feature Extraction: The corrected color information from the ROI (e.g., average R, G, B values, or values converted to another color space like LAB) is extracted and normalized to form a feature vector.
- Machine Learning-Based Prediction:
- Model Training: A Support Vector Machine (SVM) or Support Vector Regression (SVR) model is trained on a dataset of colorimetric samples with known pesticide concentrations. The model learns the nonlinear relationship between the color feature vector and the concentration [6]. The Radial Basis Function (RBF) kernel is often used for this purpose:
K(x_i, x_j) = exp(-||x_i - x_j||^2 / (2σ^2))[6]. - Inference: The trained model is converted into a lightweight format (e.g., ONNX) and embedded into the mobile app. The feature vector from the new sample is fed into this model, which outputs the predicted pesticide concentration [6]. A dual-stage approach using SVM for classification and SVR for continuous value regression can enhance precision.
- Model Training: A Support Vector Machine (SVM) or Support Vector Regression (SVR) model is trained on a dataset of colorimetric samples with known pesticide concentrations. The model learns the nonlinear relationship between the color feature vector and the concentration [6]. The Radial Basis Function (RBF) kernel is often used for this purpose:
Essential Research Reagent Solutions
The development and operation of these biosensors rely on key biological and chemical reagents that facilitate the specific recognition and signal transduction necessary for detection.
Table 2: Key Research Reagents for Smartphone-Integrated Biosensors
| Reagent / Material | Function in the Biosensing System | Example Application |
|---|---|---|
| Nanozymes (e.g., PDFeNi Foam) [47] | Mimics the activity of natural enzymes (e.g., peroxidase) to catalyze a chromogenic reaction, providing the signal for detection. | Used in a colorimetric sensor array to rapidly detect pesticides and antioxidants within 3 minutes [47]. |
| Molecularly Imprinted Polymers (MIPs) [35] | Synthetic polymers with tailor-made cavities that act as artificial antibodies, providing highly specific recognition and binding sites for target pesticide molecules. | Used as recognition elements in optical or electrochemical sensors to detect pesticides in complex matrices like tea [35]. |
| Uranium-Organic Frameworks (UOFs) [28] | Acts as a highly sensitive and selective fluorescent probe. The presence of the target analyte (pesticide) causes a change in the fluorescence intensity. | Enabled a smartphone-integrated sensor to detect pesticides and antibiotics in food samples within 10 seconds [28]. |
| Biological Recognition Elements (e.g., AChE) [6] | Enzymes like acetylcholinesterase (AChE) are inhibited by specific classes of pesticides (e.g., organophosphates), which forms the basis for the detection mechanism. | Fixed on gold nanoparticles to produce a colorimetric reaction when exposed to pesticide samples [6]. |
| Chromogenic Substrates (e.g., TMB, OPD) [47] [6] | Colorless compounds that are converted into a colored product in the presence of a catalyst (e.g., peroxidase or nanozyme), generating the measurable signal. | Oxidized by PDFeNi foam to produce distinct colors (blue, yellow, purple) for a sensor array [47]. |
The integration of hardware, software, and advanced algorithms creates a powerful and accessible platform for visual pesticide detection. The protocols outlined herein provide a framework for researchers to develop and refine these systems. Future advancements will likely focus on overcoming existing challenges, such as multi-analyte detection in even more complex matrices, further miniaturization of hardware, and the integration of more powerful AI models for data analysis. The convergence of nanomaterials, microfluidics, and artificial intelligence with smartphone technology is poised to deliver robust, lab-grade analytical capabilities directly into the hands of users, revolutionizing food safety and environmental monitoring.
The escalating global consumption of pesticides, which reached 3.7 million tons by 2022, poses significant threats to ecosystem integrity and human health through residue accumulation in agri-food products [35]. Organophosphorus pesticides (OPPs), herbicides, and fungicides represent particularly concerning classes due to their widespread use and potential toxicity, necessitating robust monitoring methodologies [49]. Conventional detection techniques like high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) and gas chromatography-mass spectrometry (GC-MS) offer excellent sensitivity but suffer from limitations including operational complexity, high costs, and lack of field-deployability [35] [49].
Smartphone-integrated biosensors have emerged as transformative analytical platforms that bridge the gap between laboratory-grade accuracy and field-based usability [35] [32]. These systems leverage smartphones' capabilities as portable, affordable analytical devices equipped with high-resolution cameras, sensors, and sophisticated operating systems [32]. This document presents detailed application notes and experimental protocols for detecting major pesticide classes using smartphone-based biosensing platforms, providing researchers with practical frameworks for implementing these advanced detection methodologies within agri-food safety monitoring systems.
Smartphone-Based Sensing Platforms: Technical Foundations
Smartphone-based detection systems typically utilize the device's complementary metal-oxide semiconductor (CMOS) or charge-coupled device (CCD) image sensors for signal acquisition [32]. These sensors employ RGB (red, green, blue) color filters with specific wavelength ranges of 600–700 nm (R), 500–600 nm (G), and 400–500 nm (B), with color intensity expressed in absolute ratios from 0-255 [32]. For enhanced analytical performance, color space transformations to HSV (hue, saturation, value) or CMYK (cyan, magenta, yellow, black) may be employed to mitigate external factors like illumination variations [32].
The integration of molecularly imprinted polymers (MIPs) as recognition elements has significantly advanced sensor capabilities, offering advantages including high selectivity, exceptional stability, cost-effectiveness, and compatibility with various transducer systems [35]. MIPs function as synthetic antibody mimics, creating template-specific cavities that enable selective recognition and binding of target pesticide molecules even within complex food matrices [35].
Table 1: Smartphone Imaging Specifications for Pesticide Detection
| Component | Specifications | Application in Pesticide Detection |
|---|---|---|
| Image Sensor | CMOS or CCD with Bayer filter array | Captures optical signals from colorimetric/fluorescent assays |
| Color Channels | RGB (R:600-700nm, G:500-600nm, B:400-500nm) | Quantitative analysis via intensity changes (0-255 scale) |
| Alternative Color Spaces | HSV, CMYK | Minimizes ambient light interference; improves accuracy |
| Data Processing | Image processing software (ImageJ, MATLAB) | Converts visual signals to quantitative data |
Case Study 1: Fluorescence Biosensor for Organophosphorus Pesticides
Principle and Signaling Pathway
This detection platform utilizes the enzymatic inhibition of alkaline phosphatase (ALP) by organophosphorus pesticides (OPs) [50]. In the absence of OPs, ALP catalyzes the hydrolysis of L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (AAP) to produce L-ascorbic acid (AA). The generated AA then reacts with o-phenylenediamine (OPD) to form the fluorescent compound 3-(1,2-dihydroxyethyl)furo[3,4-b]quinoxalin-1(3H)-one (DFQ), which exhibits strong fluorescence emission at 425 nm [50]. When OPs are present, they inhibit ALP activity, reducing AA production and consequently diminishing DFQ formation and fluorescence intensity in a concentration-dependent manner [50].
Experimental Protocol
Reagents and Materials
- Alkaline phosphatase (ALP, from bovine intestinal mucosa)
- L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (AAP)
- o-phenylenediamine (OPD)
- Organophosphorus pesticide standards (malathion as representative compound)
- Buffer solutions (phosphate buffer, pH 7.4)
- Vegetable samples (for validation studies)
- Smartphone-integrated fluorescence detection device (custom-built)
Procedure
Sample Preparation:
- Homogenize 5 g of vegetable sample with 10 mL of acetonitrile using a vortex mixer for 2 minutes.
- Centrifuge at 4000 rpm for 10 minutes and collect the supernatant.
- Filter through a 0.45 μm membrane filter prior to analysis.
Detection Procedure:
- Mix 50 μL of sample extract (or OP standard solution) with 50 μL of ALP solution (1 U/mL) in a microcentrifuge tube.
- Incubate the mixture at 37°C for 15 minutes to allow OP-mediated enzyme inhibition.
- Add 50 μL of AAP solution (5 mM) and incubate for an additional 20 minutes at 37°C.
- Introduce 50 μL of OPD solution (10 mM) and incubate for 30 minutes at 37°C in the dark to facilitate DFQ formation.
- Transfer 100 μL of the reaction mixture to a microcuvette and place in the smartphone-based fluorescence detector.
Smartphone-Based Detection:
- Position the sample in the custom portable fluorescence device equipped with an appropriate excitation source (365 nm UV LED) and emission filter (430 nm longpass).
- Capture the fluorescence image using the smartphone camera through the device optics.
- Utilize a dedicated smartphone application to convert the fluorescence intensity to RGB values.
- Correlate the blue channel intensity with OP concentration based on a pre-established calibration curve.
Data Analysis:
- Generate a calibration curve using malathion standards (0.05-1.0 ppm).
- Calculate OP concentration in samples from the linear regression equation.
- Validate method accuracy by comparison with HPLC (for vegetable samples).
Performance Metrics
This fluorescence biosensor demonstrates a detection limit of 0.05 ppm for malathion with a linear range of 0.1-1.0 ppm [50]. The smartphone-based detection system shows approximately 70 times higher sensitivity compared to conventional spectrofluorometers, enabling precise quantification at trace levels [50]. Method validation with vegetable samples shows excellent agreement with standard HPLC methodologies, confirming practical applicability for food safety monitoring [50].
Case Study 2: Molecularly Imprinted Polymer-Based Sensor for Herbicides
Principle and Experimental Workflow
Molecularly imprinted polymers (MIPs) serve as synthetic recognition elements created by polymerizing functional monomers around a template herbicide molecule [35]. After template removal, complementary binding cavities remain that selectively rebind the target herbicide from complex matrices [35]. When integrated with smartphone detection, these platforms typically employ colorimetric or fluorescence transduction mechanisms for herbicide quantification.
Experimental Protocol
MIP Synthesis and Characterization
Herbicide-Imprinted Polymer Synthesis:
- Dissolve the template herbicide (e.g., glyphosate, atrazine) and functional monomer (e.g., methacrylic acid) in a porogenic solvent (acetonitrile or toluene) at molar ratio of 1:4.
- Add cross-linker (ethylene glycol dimethacrylate) and initiator (azobisisobutyronitrile, AIBN).
- Purge with nitrogen gas for 5 minutes to remove oxygen.
- Polymerize under UV irradiation (365 nm) for 24 hours at room temperature.
- Grind the resulting polymer blocks and sieve to obtain particles of 25-50 μm diameter.
Template Removal:
- Extract template molecules by Soxhlet extraction with methanol:acetic acid (9:1, v/v) for 24 hours.
- Dry the extracted MIPs under vacuum at 60°C for 12 hours.
- Confirm template removal by HPLC analysis of extraction solvents.
MIP Characterization:
- Analyze binding capacity through batch adsorption experiments.
- Evaluate selectivity against structurally similar compounds.
- Characterize morphology by scanning electron microscopy.
Sensor Assembly and Detection
Sensor Preparation:
- Immobilize MIP particles on a glass substrate or incorporate into a paper-based sensor strip.
- For optical detection, incorporate signal probes (gold nanoparticles, quantum dots, or chromogenic reagents).
Herbicide Detection Protocol:
- Incubate the MIP sensor with sample solution (or standard) for 15-20 minutes.
- Wash with buffer to remove non-specifically bound compounds.
- For colorimetric detection, measure color intensity changes using smartphone RGB analysis.
- For fluorescent MIPs, capture fluorescence emission using a smartphone-based fluorescence detector.
Smartphone Quantification:
- Capture sensor image under controlled lighting conditions.
- Extract RGB values using image processing applications (ImageJ, MATLAB, or custom apps).
- Calculate herbicide concentration from calibration curves.
Performance Metrics
MIP-based sensors exhibit exceptional selectivity for target herbicides with cross-reactivity below 15% for most structurally related compounds [35]. Detection limits typically range from 0.01-0.1 ppm, satisfying regulatory requirements for maximum residue limits (MRLs) in food commodities [35]. The sensors demonstrate excellent stability, maintaining recognition capability after more than 50 regeneration cycles [35].
Table 2: Performance Comparison of Smartphone-Based Pesticide Detection Methods
| Detection Platform | Target Pesticides | Linear Range | Detection Limit | Sample Matrix |
|---|---|---|---|---|
| Fluorescence Biosensor (ALP-OPD) [50] | Organophosphates (e.g., malathion) | 0.1–1.0 ppm | 0.05 ppm | Vegetable samples |
| Molecularly Imprinted Polymers [35] | Herbicides, fungicides, OPs | 0.01–10 ppm | 0.001–0.01 ppm | Complex food matrices |
| Smartphone Colorimetry [32] | Multiple classes | Varies by assay | Varies by assay | Agricultural products |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Reagents for Smartphone-Based Pesticide Detection
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Alkaline Phosphatase (ALP) | Enzyme inhibition target for OP detection | Fluorescence biosensor for organophosphates [50] |
| L-ascorbic acid 2-phosphate (AAP) | Enzyme substrate for ALP | Converted to ascorbic acid in OP detection assay [50] |
| o-Phenylenediamine (OPD) | Fluorogenic probe | Reacts with ascorbic acid to form fluorescent DFQ [50] |
| Molecularly Imprinted Polymers | Synthetic recognition elements | Selective herbicide/fungicide capture in sensors [35] |
| Methacrylic Acid | Functional monomer for MIP synthesis | Creates complementary binding sites in polymer matrix [35] |
| Ethylene Glycol Dimethacrylate | Cross-linking agent for MIP synthesis | Provides structural stability to imprinted polymers [35] |
| Gold Nanoparticles | Colorimetric probes | Signal generation in colorimetric pesticide sensors [32] |
| Smartphone Image Analysis Apps | Signal processing and quantification | Convert visual data to concentration values (ImageJ, MATLAB) [32] |
Smartphone-integrated biosensors represent a paradigm shift in pesticide detection technology, offering unprecedented opportunities for decentralized food safety monitoring. The case studies presented herein demonstrate robust methodologies for detecting organophosphates, herbicides, and fungicides in agri-food products with sensitivity comparable to conventional laboratory techniques [35] [50].
Future developments in this field will likely focus on multiplexed detection platforms for simultaneous screening of multiple pesticide residues, enhanced by machine learning algorithms for data interpretation [35] [32]. The integration of smartphone-based sensors with cloud computing and IoT technologies will facilitate real-time monitoring and data sharing across the food supply chain, ultimately strengthening global food safety systems [32] [8]. As these technologies mature, they hold tremendous potential to transform regulatory compliance monitoring, enabling more frequent and comprehensive pesticide surveillance while reducing analytical costs and time-to-result.
Overcoming Deployment Hurdles: Sensor Calibration, Interference, and Robustness
Addressing Sensor Performance Variability and Calibration Inconsistencies
Smartphone-integrated biosensors represent a transformative technology for the visual detection of pesticide residues, offering the potential for rapid, on-site analysis. However, the widespread adoption and reliability of these systems are fundamentally challenged by sensor performance variability and calibration inconsistencies. These issues arise from the complex interplay of biological recognition elements, transducer stability, and the variable operational conditions encountered in field use. This document provides detailed application notes and protocols to characterize, mitigate, and manage these variability sources, ensuring the generation of precise and reproducible data for researchers and drug development professionals.
Performance Variability: Challenges and Quantitative Characterization
The performance of smartphone-based biosensors is influenced by multiple factors, leading to potential inaccuracies if not properly managed. The key challenges and their quantitative manifestations are summarized below.
Table 1: Key Challenges in Smartphone-Integrated Biosensor Performance
| Challenge Category | Specific Source of Variability | Impact on Sensor Performance |
|---|---|---|
| Fundamental Sensor Physics | Low Signal-to-Noise Ratio (SNR) at ultralow concentrations [51] | Faint analyte signals are obscured by electronic noise, complicating distinction from false positives [51]. |
| Sensor Selectivity and Cross-Interference [51] [1] | Non-target molecules (e.g., other pesticides, tea polyphenols) trigger a response, leading to overestimation [1]. | |
| Environmental Stressors | Temperature Fluctuations [51] [52] | Causes physical expansion/contraction of sensor materials and electronic variability, inducing calibration drift [52]. |
| Humidity Variations [52] | High humidity can cause condensation and corrosion; low humidity can desiccate sensitive elements [52]. | |
| Particulate Accumulation (Dust) [52] | Obstructs sensor surfaces, altering exposure to the sample and skewing readings [52]. | |
| System Integration & Use | Biological Recognition Element Stability [1] [35] | Enzymes (e.g., AChE) can be inactivated by temperature, pH, or reactive oxygen species [35]. |
| Smartphone Camera & Illumination Variance [6] | Differences in camera sensors, auto-white balance, and ambient lighting affect colorimetric data fidelity [6]. |
The quantitative impact of these variability sources can be characterized through key performance metrics, as outlined in the following table.
Table 2: Quantitative Performance Metrics and Targets for Smartphone-Based Biosensors
| Performance Metric | Description | Typical Target for Reliable Field Use |
|---|---|---|
| Detection Limit | The lowest concentration of an analyte that can be reliably distinguished from zero [1]. | Parts-per-billion (ppb) to parts-per-trillion (ppt) range for pesticides [51]. |
| Calibration Drift | The deviation in sensor output over time under constant conditions, often expressed as signal loss per day [52] [53]. | Minimized via regular calibration; influenced by environmental stressors [52]. |
| Signal-to-Noise Ratio (SNR) | The ratio of the power of the true analyte signal to the power of the background noise [51]. | Maximized using signal processing (e.g., averaging) and low-noise design [51]. |
| Analysis Time | The time required from sample introduction to result output [1]. | 5 to 30 minutes for biosensors, enabling rapid on-site screening [1]. |
Experimental Protocols for Characterization and Mitigation
Protocol 1: Assessing Environmental Stressor-Induced Drift
Objective: To quantify the impact of temperature and humidity on the calibration stability of the smartphone-based colorimetric biosensor.
Materials:
- Smartphone biosensor platform with a standardized colorimetric test strip.
- Environmental chamber (or controlled temperature/humidity boxes).
- Standard pesticide solutions at known concentrations (e.g., 0 ppb, 10 ppb, 50 ppb).
- Data logging equipment.
Methodology:
- Initial Calibration: At a stable room temperature (e.g., 25°C) and humidity (50% RH), capture images of test strips exposed to the standard solutions. Build the initial calibration model (e.g., using SVR) [6].
- Stress Exposure: Place the biosensor test strips in the environmental chamber. Subject them to a matrix of conditions:
- Temperature: 4°C, 25°C, 40°C (at constant 50% RH).
- Humidity: 30% RH, 50% RH, 80% RH (at constant 25°C).
- Exposure Time: Maintain each condition for 1, 4, and 24 hours.
- Data Collection: After each time interval, use the smartphone app to capture images of the test strips and record the predicted concentration.
- Data Analysis: Calculate the percentage drift from the known concentration for each condition. Plot drift against temperature, humidity, and time to establish sensor-specific tolerance thresholds.
Protocol 2: Validating Cross-Interference and Selectivity
Objective: To determine the sensor's specificity towards the target pesticide in the presence of common interferents found in complex matrices like tea.
Materials:
- Smartphone biosensor platform.
- Target pesticide standard (e.g., imidacloprid).
- Common interferents: other pesticides (e.g., pyrethroids), tea polyphenols, caffeine, and heavy metal ions.
- Purified water or a simulated tea matrix.
Methodology:
- Preparation: Prepare solutions containing:
- The target pesticide alone at its Maximum Residual Limit (MRL).
- Each potential interferent alone at a concentration typical for tea.
- A mixture of the target pesticide and each interferent.
- A mixture of all interferents without the target pesticide.
- Measurement: Apply each solution to the test strip and perform the detection protocol using the smartphone app. Record the measured concentration.
- Analysis: A sensor with high selectivity will show a significant signal only for solutions containing the target pesticide. The signal from mixtures should not statistically differ from the target-alone signal. A false positive is indicated by a signal from an interferent-alone solution.
Protocol 3: Standardized Workflow for Robust Calibration
Objective: To establish a detailed procedure for calibrating the smartphone-biosensor system, integrating illumination correction to minimize device-to-device variability.
Materials:
- Smartphone with the dedicated application installed [6].
- Reference color card (e.g., with neutral gray, white, and primary color patches).
- Batch of colorimetric test strips from the same production lot.
- Series of standard pesticide solutions with concentrations spanning the dynamic range.
Methodology:
- System Setup: Launch the application and navigate to the calibration module.
- Illumination Correction:
- Position the reference color card within the camera's view, under the same lighting conditions used for sample testing.
- The application will automatically capture an image and calculate a 3x3 color conversion matrix. This matrix maps the captured colors under ambient light to their known reference values, correcting for lighting variations [6].
- Save these correction parameters for future use with samples tested under the same lighting.
- Building the Calibration Curve:
- For each standard solution, apply a droplet to a test strip and allow the colorimetric reaction to proceed for the prescribed time.
- Capture an image of the strip within the app's guided overlay.
- The app will automatically perform region selection and apply the pre-determined illumination correction.
- Extract the color feature vector (e.g., RGB, LAB values) from the reactive zone.
- Repeat in triplicate for each standard.
- Model Training: Input the known concentrations and the averaged color feature vectors into the machine learning model (SVM/SVR) to train the calibration model [6]. The model is then exported (e.g., as an ONNX file) and embedded into the application for future predictions.
Figure 1: Workflow for standardized system calibration.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for Biosensor Development
| Item | Function/Description | Application in Protocol |
|---|---|---|
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymers with cavities complementary to a target pesticide, serving as stable, artificial recognition elements [35]. | Used as the sensing interface to selectively capture target molecules, reducing cross-interference [35]. |
| Enzymes (e.g., Acetylcholinesterase - AChE) | Biological recognition element. Pesticides like organophosphates inhibit AChE activity, which is transduced into a measurable signal [1] [6]. | Core component of enzymatic biosensors; its inhibition is correlated with pesticide concentration [6]. |
| Gold Nanoparticles (AuNPs) | Nanomaterial used as a colorimetric label or to enhance signal transduction. Their aggregation or color change indicates analyte presence [6]. | Used in test strips; color change from reaction provides the signal for smartphone detection [6]. |
| Smartphone Application with ML | Cross-platform app (e.g., built with Flutter) that handles image capture, correction, and concentration prediction [6]. | Executes the entire analysis workflow, from image acquisition to result visualization, ensuring user-independent results [6]. |
| Reference Color Card | A physical card with patches of known, stable colors. | Serves as a reference for the software to perform automatic illumination correction, mitigating lighting variability [6]. |
Data Analysis and Computational Workflow
The data processing pipeline within the smartphone application is critical for overcoming inherent variability. The following diagram and description detail this workflow.
Figure 2: Smartphone app data processing and analysis workflow.
Workflow Description:
- Image Capture & Preprocessing: The application guides the user to capture an image of the test strip. It then automatically selects the region of interest (ROI) and applies illumination correction algorithms, such as the Gray World method or color card-based transformation, to normalize the image colors [6].
- Feature Extraction: The mean color values from the ROI are extracted, typically in RGB or a more perceptually uniform color space like LAB, forming a feature vector.
- Machine Learning Prediction: The normalized color vector is fed into a pre-trained, dual-stage machine learning model.
- Result Fusion: The final reported concentration is a fusion of the SVM and SVR outputs, often constrained to the range of the predicted class to prevent over- or under-prediction, ensuring a robust and accurate result [6].
Addressing sensor performance variability is not merely a preliminary step but a continuous requirement throughout the lifecycle of a smartphone-integrated biosensor. By systematically applying the protocols for environmental testing, selectivity validation, and standardized calibration outlined in this document, researchers can significantly enhance the reliability of their data. The integration of robust materials like MIPs, coupled with a computational workflow that leverages machine learning and illumination correction, provides a powerful strategy to overcome inherent inconsistencies. This structured approach to managing variability is foundational to developing pesticide detection systems that are not only innovative but also dependable for critical decision-making in food safety and environmental health.
Mitigating Environmental and Matrix Interferences in Complex Samples
The transition of biosensors from controlled laboratory settings to on-field applications for pesticide detection introduces significant challenges related to environmental and matrix interferences. Complex samples such as fruits, vegetables, and environmental water contain diverse interfering substances including pigments, proteins, carbohydrates, lipids, and inorganic salts that can compromise detection accuracy through non-specific binding, optical interference, or sensor fouling [54] [55]. For smartphone-integrated visual biosensors, these interferences manifest as false positives/negatives, reduced sensitivity, and impaired quantitative capability, ultimately limiting their practical utility in real-world scenarios. This document outlines systematic strategies and detailed protocols to mitigate these challenges, enabling reliable pesticide detection in complex matrices.
Interference Challenges and Mitigation Strategies
Table 1: Common Interference Sources and Corresponding Mitigation Approaches in Visual Pesticide Detection
| Interference Category | Specific Challenges | Mitigation Strategies | Key Materials & Technologies |
|---|---|---|---|
| Optical Interferences | Sample turbidity; endogenous chromophores; light scattering [54] | Ratiometric sensing; sample filtration; plasmonic nanomaterials [56] [57] | Metal-organic frameworks (MOFs); quantum dots; gold nanoparticles |
| Matrix Effects | Non-specific protein adsorption; enzymatic inhibitors; pH variations [42] [58] | Sample dilution; enzyme immobilization; nanofiber carriers [42] [58] | Crosslinked PVA/CA nanofiber mats; acetylcholinesterase (AChE) |
| Environmental Factors | Temperature fluctuations; humidity; heterogeneous distribution [55] | Internal referencing; smartphone temperature logging; controlled incubation [57] [55] | Distance-readout ECL; smartphone environmental sensors |
| Cross-Reactivity | Structural analogs; metabolites; coexisting contaminants [54] | Aptamer-based recognition; molecularly imprinted polymers (MIPs) [57] [55] | DDVP-specific aptamers; synthetic MIPs |
Material and Methods
Research Reagent Solutions
Table 2: Essential Materials for Interference-Mitigated Visual Biosensing
| Material / Reagent | Function / Application | Specific Role in Interference Mitigation |
|---|---|---|
| Polyvinyl Alcohol/Citric Acid (PVA/CA) Nanofiber Mat | Enzyme immobilization matrix [42] [58] | Provides high surface area, water stability, and reduced swelling in aqueous samples |
| EDC/NHS (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide) | Carboxyl group activation for surface decoration [42] [58] | Enhances enzyme loading capacity and stability via covalent immobilization |
| Acetylcholinesterase (AChE) | Enzyme inhibition-based detection [10] [42] | Biological recognition element for organophosphate pesticides |
| p-Fe₃O₄@PDA@ZIF-8 Nanozyme | Peroxidase-like catalyst in ECL sensors [57] | Enables equipment-free distance readout, resistant to environmental interference |
| Chitosan/AChE/PAnNF/CNT Nanocomposite | Conductance-based sensing film [10] | Provides anti-interference properties through selective doping mechanisms |
| Organophosphate-specific Aptamers | Molecular recognition elements [57] | High specificity reduces cross-reactivity with non-target compounds |
Experimental Protocol: Nanofiber Mat-Based Visual Detection Card
Fabrication of Crosslinked PVA/CA Nanofiber Mat (PCNM)
Reagents: Polyvinyl alcohol (PVA), Citric Acid (CA), Deionized water [42] [58]
Procedure:
- Prepare PVA solutions (5%, 7.5%, 10%, 12.5% w/v) by dissolving in deionized water at 90°C with continuous stirring for 4 hours
- Incorporate CA into PVA solutions at 20% (w/w relative to PVA weight) with additional stirring for 2 hours
- Characterize solution properties: measure conductivity using DDS-307A conductivity meter and viscosity with LV-SSR viscometer
- Electrospinning parameters:
- Voltage: 13-17 kV
- Flow rate: 0.25-0.65 mL/h
- Collector distance: 11-15 cm
- Environmental conditions: 25±2°C, 50±2% relative humidity
- Collection time: 10 hours
- Thermal crosslinking:
- Temperature: 130°C
- Duration: 50 minutes
- Outcome: Appropriate microstructure and water stability for AChE immobilization
Surface Decoration and Enzyme Immobilization
Reagents: EDC/NHS, Acetylcholinesterase (AChE), Phosphate Buffered Saline (PBS, pH 7.4) [42] [58]
Procedure:
- Cut crosslinked PCNM into 1 cm diameter discs
- Surface decoration:
- Immerse PCNM discs in 1 M EDC/NHS PBS solution
- Incubate at 4°C for 1-4 hours
- Rinse with PBS to remove excess EDC/NHS
- Enzyme immobilization:
- Incubate decorated nanofiber mats (E-PCNM) in 1 mg/mL AChE PBS solution (pH 7.4)
- Maintain at 4°C for 1.5 hours
- Result: Enzyme card (EC) ready for use
- Quality control:
- Assess enzyme loading using BCA protein assay kit
- Verify enzyme secondary structure integrity via circular dichroism (CD) measurements
Detection Procedure for Complex Samples
Reagents: Substrate card (SC) with indoxyl acetate (IA), Food samples (fruits/vegetables), PBS buffer [42] [58]
Procedure:
- Sample preparation:
- Homogenize food samples (1 g) in PBS (10 mL)
- Centrifuge at 5000 rpm for 5 minutes
- Collect supernatant for analysis
- Inhibition step:
- Incubate enzyme card (EC) with 100 μL sample supernatant for 10 minutes
- Pesticides present in sample inhibit AChE activity
- Color development:
- Combine inhibited EC with substrate card (SC) containing 3 mg/mL IA
- Incubate for 1 minute
- Visual observation: Reduced color development indicates pesticide presence
- Smartphone quantification:
- Capture image using smartphone camera under standardized lighting
- Analyze color intensity using image processing applications
- Compare with calibration standards for quantification
Signaling Pathways and Experimental Workflows
Visual Detection Workflow: Diagram illustrating the complete process from nanofiber mat preparation to smartphone-based result analysis.
Interference Mitigation Mechanism: Diagram showing the biosensing principle and pesticide inhibition pathway for conductance-based detection.
Performance Metrics and Validation
Table 3: Quantitative Performance of Interference-Mitigated Biosensing Platforms
| Platform & Detection Principle | Target Pesticide | Linear Range | Detection Limit | Interference Resistance | Reference Matrix |
|---|---|---|---|---|---|
| Smartphone/Resistive Biosensor [10] | Paraoxon-Methyl | 1 ppt - 100 ppb | 0.304 ppt | RSD <5%; 98.3% recovery rate | Food/Water samples |
| Visual Nanofiber Card [42] [58] | Phoxim | - | 0.007 mg/L | 11 min detection; superior to commercial cards | Fruit/Vegetable samples |
| Visual Nanofiber Card [42] [58] | Methomyl | - | 0.10 mg/L | Reduced hydrophilicity; minimized swelling | Fruit/Vegetable samples |
| Distance-Readout ECL Sensor [57] | Dichlorvos (DDVP) | 50-1200 pM | - | Equipment-free; resistant to intensity variations | Vegetable samples |
| AChE/Chitosan/PANiNF/CNT [10] | Organophosphates | Wide dynamic range | High sensitivity | Minimal sample requirement; integrated components | Environmental Water |
The integration of advanced materials such as crosslinked nanofiber mats, specific biorecognition elements, and smartphone-based readout systems provides a robust framework for mitigating environmental and matrix interferences in complex samples. The protocols outlined herein enable researchers to achieve reliable, sensitive, and specific detection of pesticide residues in field conditions, advancing the applicability of smartphone-integrated biosensors for real-world food safety and environmental monitoring applications.
Strategies for Enhancing Selectivity and Minimizing False Positives
The integration of biosensors with smartphones has revolutionized the on-site detection of pesticides, offering a powerful tool for ensuring agricultural and food safety. However, the practical deployment of these systems is often challenged by issues of selectivity and false positives, particularly when dealing with complex sample matrices like tea, fruits, and vegetables. False positives can arise from non-specific binding, matrix interference, cross-reactivity of biological recognition elements, or environmental factors affecting the sensor platform. This application note details targeted strategies and validated experimental protocols to enhance the selectivity and reliability of smartphone-integrated visual biosensors for pesticide detection. We provide actionable methodologies focusing on material design, surface functionalization, data processing, and system integration to minimize erroneous results and ensure accurate, field-deployable analysis for researchers and scientists.
Smartphone-based biosensors leverage the device's high-resolution camera, processing power, and connectivity to function as portable, user-friendly analytical systems [32]. These platforms often utilize colorimetric, fluorescent, or electrochemical biosensing mechanisms, where the smartphone camera captures visual changes induced by the target analyte. A significant challenge is that constituents of complex agricultural samples, such as polyphenols and alkaloids in tea, can cause non-specific signals, leading to false positives [1]. Enhancing selectivity requires a multi-faceted approach, from the initial design of the biorecognition element to the final data analysis algorithm. This document outlines a comprehensive set of strategies and provides detailed protocols to systematically address these challenges, thereby improving the robustness of analytical results.
Strategic Approaches and Underlying Principles
Optimizing the components and data processing workflow of the biosensor is fundamental to achieving high selectivity. The following strategies are critical for minimizing false positives.
Advanced Biorecognition Elements
The choice of the biorecognition molecule is the first line of defense against false positives.
- Aptamers: Synthetic single-stranded DNA or RNA oligonucleotides selected for high affinity to specific targets offer significant advantages over traditional antibodies. Their synthetic nature allows for precise chemical modification and regeneration. Furthermore, they can be selected under controlled conditions that mimic the sample matrix, thereby improving specificity and reducing non-specific binding from the outset.
- Molecularly Imprinted Polymers (MIPs): These artificial antibodies are synthetic polymers with cavities complementary to the target molecule in shape, size, and functional groups. MIPs exhibit excellent physical and chemical stability, are resistant to harsh environments, and can be engineered for specific pesticides, drastically reducing cross-reactivity with structurally similar compounds.
- Enzyme Inhibition-Based Sensing: This approach exploits the specific inhibition of enzymes like acetylcholinesterase (AChE) by organophosphorus and carbamate pesticides [1]. The selectivity is inherent to the enzyme-inhibitor interaction. Using purified enzymes with high specific activity is crucial to minimize interference from other sample components.
Nanomaterial-Enhanced Specificity and Signal Fidelity
Nanomaterials are not merely signal amplifiers; they can be engineered to improve selectivity.
- High-Fidelity Signal Probes: The use of nanomaterials like gold-silver nanostars for Surface-Enhanced Raman Scattering (SERS) provides a unique vibrational "fingerprint" for the target molecule [26]. This fingerprinting capability directly counters false positives by confirming the identity of the adsorbed molecule, not just its presence.
- Matrix Blocking and Selective Filtering: Nanomaterials such as polydopamine coatings and metal-organic frameworks (MOFs) can be functionalized to act as a selective filter [26]. They can be designed to pre-concentrate the target analyte while physically blocking or repelling larger interfering molecules like proteins or colored pigments present in food samples.
Smartphone-Enhanced Data Acquisition and Processing
The smartphone itself can be leveraged to transcend the limitations of subjective visual inspection.
- Multi-Parameter Color Analysis: Instead of relying on a single color channel (e.g., Red, Green, Blue or RGB), advanced image processing algorithms can extract multiple color space values (e.g., Hue, Saturation, Value or HSV; Cyan, Magenta, Yellow, Black or CMYK) from the region of interest [32]. This multi-dimensional data provides a more robust signature of the positive signal, making it easier to distinguish from non-specific background color changes.
- AI and Machine Learning-Based Classification: Artificial intelligence (AI) can be trained to recognize complex patterns in the sensor's output data that are indicative of a true positive. By training a classifier with data from known positive samples, negative controls, and samples with common interferents, the system can learn to accurately identify and reject false positive signals [32]. This is particularly powerful when combined with multi-parameter color data.
Table 1: Comparison of Biorecognition Elements for Selectivity
| Element Type | Mechanism of Selectivity | Advantages for Minimizing False Positives | Common Challenges |
|---|---|---|---|
| Aptamers | Folding into 3D structures with high-affinity binding pockets | Can be selected against specific interferents; minimal batch-to-batch variation | Susceptible to nuclease degradation in complex samples |
| Molecularly Imprinted Polymers (MIPs) | Shape-complementary cavities and chemical interactions | High chemical stability; resistant to denaturation | Risk of heterogeneous binding sites leading to cross-reactivity |
| Enzymes (e.g., AChE) | Specific catalytic site inhibition by target pesticides | Well-characterized inhibition kinetics; broad-detection class | Can be inhibited by other compounds (e.g., heavy metals) |
Experimental Protocols
The following protocols provide a step-by-step guide for key experiments aimed at validating selectivity and minimizing false positives.
Protocol: Selectivity Profiling of a Biosensor Using Common Interferents
Objective: To experimentally verify that the biosensor produces a negligible response to substances commonly found in the target sample matrix, thereby confirming high selectivity.
Materials:
- Smartphone-integrated biosensor platform
- Stock solution of target pesticide (e.g., 100 ppm)
- Stock solutions of potential interferents (e.g., other pesticides with similar structures, common ions (Ca²⁺, Mg²⁺), sugars, organic acids, humic acid)
- Sample matrix blank (e.g., buffer or extracted sample without pesticides)
- Micropipettes and sterile tips
Procedure:
- Preparation: Dilute the stock solution of the target pesticide to a concentration near the sensor's limit of detection (LOD). Prepare solutions of each potential interferent at a concentration 10 times higher than that expected in the real sample.
- Baseline Measurement: Analyze the sample matrix blank using the standard biosensor assay protocol. Record the output signal (e.g., RGB values, electrochemical current). This is your baseline (negative control).
- Target Analysis: Analyze the diluted target pesticide solution. Record the signal. This is your positive control.
- Interferent Analysis: Individually analyze each solution of potential interferents using the exact same protocol.
- Cross-Reactivity Calculation: Calculate the signal response for each interferent relative to the target response using the formula: Cross-Reactivity (%) = (Signal_Interferent - Signal_Blank) / (Signal_Target - Signal_Blank) × 100%
- Acceptance Criterion: A well-designed sensor should typically show cross-reactivity of <5% for most non-target interferents.
Protocol: Optimization of Surface Blocking Agents to Reduce Non-Specific Binding
Objective: To identify the most effective blocking agent that minimizes non-specific adsorption of matrix components onto the sensor surface without inhibiting the specific recognition event.
Materials:
- Functionalized sensor substrates (e.g., aptamer-conjugated gold nanoparticles, antibody-immobilized electrodes)
- Candidate blocking agents (e.g., 1-5% Bovine Serum Albumin (BSA), 1% casein, 0.1% Tween-20, commercial protein-based blockers)
- Spiked sample solution (target pesticide in a cleaned sample matrix)
- Negative control (sample matrix without pesticide)
- Washing buffers (e.g., Phosphate Buffered Saline with Tween-20, PBST)
Procedure:
- Surface Preparation: Divide the functionalized sensor substrates into several groups, one for each blocking agent to be tested, plus an unblocked control.
- Blocking: Incubate each group with a different blocking solution for 30-60 minutes at room temperature with gentle shaking.
- Washing: Gently wash all sensors three times with washing buffer to remove unbound blocking agent.
- Sample Exposure: Apply the spiked sample solution to one sensor from each blocked group and the unblocked control. Apply the negative control to another sensor from each group.
- Signal Detection: Perform the standard signal detection procedure using the smartphone platform.
- Analysis: Calculate the signal-to-noise ratio (SNR) for each blocking condition. SNR = (Signal_Spiked - Signal_Negative) / Standard Deviation_Negative
- Selection: The blocking agent that yields the highest SNR is the most effective, as it maximizes the specific signal while minimizing the non-specific background (noise).
Research Reagent Solutions
The following table details key materials essential for constructing selective and robust smartphone-integrated biosensors.
Table 2: Essential Research Reagents and Materials
| Reagent/Material | Function/Application in Biosensor |
|---|---|
| Nucleic Acid Aptamers | Synthetic biorecognition elements; selected for high specificity to target pesticides like acetamiprid or ochratoxin [26]. |
| Gold Nanoparticles (AuNPs) & Au-Ag Nanostars | Colorimetric signal probes (color change from red to blue upon aggregation) or as a platform for SERS-based detection, providing unique analyte fingerprints [26]. |
| Polydopamine & Melanin-like Materials | Used for versatile surface coating and functionalization; improves biocompatibility and can be used to modify electrodes in electrochemical sensors [26]. |
| Bovine Serum Albumin (BSA) | A common blocking agent used to passivate unused binding sites on the sensor surface, thereby reducing non-specific adsorption of interferents. |
| Carbodiimide Crosslinkers (e.g., EDC) | Used for covalent immobilization of biorecognition elements (e.g., antibodies, aptamers) onto sensor surfaces via carboxyl-amine coupling [26]. |
| Quorum Sensing Molecules (AHLs) | While used in microbial biosensors, they illustrate the principle of specific biological recognition that can be adapted for chemical sensing [59]. |
Workflow and System Diagrams
The following diagrams illustrate the key workflows and logical relationships for enhancing selectivity.
Sensor Optimization Workflow
Selectivity Verification Logic
Optimizing Assay Workflow for User-Friendliness and Minimal Operational Error
This application note provides detailed protocols for developing robust and user-friendly workflows for smartphone-integrated biosensors, specifically for the visual detection of pesticide residues. By focusing on the optimization of key operational steps—from sample preparation to data interpretation—these guidelines aim to minimize procedural errors and enhance the reliability of on-site analyses. The methodologies are framed within the context of advancing field-deployable diagnostic tools for researchers and scientists in food safety and agricultural monitoring.
Smartphone-based biosensors represent a transformative approach in analytical science, merging portability with the powerful processing and imaging capabilities of mobile devices for on-site detection [60]. Their application in monitoring pesticide residues, such as organophosphorus compounds and neonicotinoids, is particularly valuable for ensuring food safety and environmental health [1] [61]. Traditional detection methods like gas chromatography (GC) or high-performance liquid chromatography (HPLC), while highly accurate, require intricate sample pretreatment, sophisticated laboratory settings, and trained personnel, making them unsuitable for rapid, field-deployable screening [1] [35].
Transitioning these analyses from the lab to the field necessitates assay workflows that are not only sensitive and specific but also intrinsically designed for user-friendliness and minimal operational error. This document outlines optimized protocols and provides structured data and visual workflows to guide researchers in implementing these advanced biosensing platforms effectively.
Summarized Quantitative Data of Biosensing Platforms
The performance of a biosensor is quantified by its sensitivity, detection limit, and speed. The table below summarizes the analytical performance of various biosensor types used for pesticide detection, as reported in recent literature. This data aids in selecting an appropriate sensing platform for specific application requirements.
Table 1: Analytical Performance of Selected Biosensors for Pesticide Detection
| Biosensor Platform | Recognition Element | Target Pesticide(s) | Limit of Detection (LOD) | Assay Time | Key Advantage |
|---|---|---|---|---|---|
| Fluorescent Microfluidic Sensor [61] | Enzyme (AChE) | Organophosphorus (OPs) | 0.38 pM | ~10 minutes | Ultra-high sensitivity |
| Paper-based Colorimetric Sensor [61] | Nanozyme (CuONPs) | Malathion (OP) | 0.08 mg/L | ~10 minutes | Cost-effective, portable |
| Molecularly Imprinted Polymer (MIP) Optical Sensor [35] | Biomimetic Polymer | Various | nM range | 5-30 minutes [1] | High stability & specificity |
| Dual-State Emissive Probe (TT1) [62] | Small Organic Molecule | Trifluralin, Fenitrothion | 180 nM | Rapid, visual | Direct visual detection on surfaces |
| Electrochemical Biosensor [1] | Aptamer/Enzyme | Various | nM to pM [1] | Rapid | Portability, high sensitivity |
The Scientist's Toolkit: Essential Research Reagents and Materials
A successful assay relies on its core components. The following table details key reagents and their functions, which are fundamental to developing and operating smartphone-integrated biosensors for visual pesticide detection.
Table 2: Key Research Reagent Solutions for Biosensor Assay Development
| Item | Function and Description in the Assay |
|---|---|
| Biological Recognition Elements | |
| Acetylcholinesterase (AChE) | An enzyme whose activity is inhibited by organophosphorus and carbamate pesticides, serving as the basis for the detection mechanism [61]. |
| Aptamers | Single-stranded DNA or RNA oligonucleotides that bind to specific pesticide targets with high affinity; offer high stability and are synthetically produced [61]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic, biomimetic polymers with custom-designed cavities that selectively recognize and bind to target pesticide molecules [35] [61]. |
| Signal Transduction Components | |
| Quantum Dots (QDs) / Fluorophores | Nanoscale semiconductors or fluorescent molecules (e.g., TPA-based emitters) that emit light at specific wavelengths upon excitation; signal changes (quenching/enhancement) indicate analyte presence [61] [62]. |
| Nanozymes (e.g., CuO NPs) | Nanomaterials that mimic the catalytic activity of natural enzymes (e.g., peroxidase), used to generate a colored product for colorimetric detection [61]. |
| Assay Substrate & Platform | |
| Paper-based Microfluidic Device | A low-cost, portable substrate that wicks samples and reagents via capillary action, facilitating simple, pump-free fluidic operations [61]. |
| Smartphone with Camera & App | The core detection and processing unit. The camera captures colorimetric or fluorescent signals, and a dedicated app processes the image for quantitative analysis [60] [35]. |
| Supporting Reagents | |
| Acetylthiocholine (ATCh) / H₂O₂ | Enzyme substrates. ATCh is hydrolyzed by AChE, producing thiocholine, which interacts with signal probes. H₂O₂ is a substrate for peroxidase-like nanozymes [61]. |
| Chromogenic Agents (e.g., TMB) | Colorless compounds that, upon oxidation (e.g., by nanozymes), produce a intense blue or other colored product, enabling visual and colorimetric readout [61]. |
Experimental Protocols
Protocol 1: Paper-based Colorimetric Assay for Organophosphorus Pesticides
This protocol details the steps for a nanozyme-based assay, ideal for field use due to its minimal steps and visual readout.
Workflow Diagram: Paper-Based Assay
Materials:
- Paper-based analytical device impregnated with acetylcholinesterase (AChE) and copper oxide nanozymes (CuONPs) [61].
- Acetylthiocholine (ATCh) iodide solution.
- Sample extract from fruits, vegetables, or tea leaves.
- Smartphone with a colorimetric analysis application installed.
Procedure:
- Sample Preparation: Homogenize the food sample (e.g., 1 g of tea leaves) and extract pesticides using a suitable solvent (e.g., acetonitrile). Filter or dilute the extract as necessary [1] [61].
- Assay Initiation: Apply a fixed volume (e.g., 50 µL) of the sample extract to the sample zone of the paper device.
- Reaction Incubation: Allow the device to incubate at room temperature for approximately 10 minutes. In the absence of pesticides, AChE will hydrolyze ATCh to produce thiocholine and H₂O₂. The CuONP nanozymes will then catalyze the oxidation of a chromogen (e.g., TMB) by H₂O₂, producing a blue color. Pesticides inhibit AChE, reducing H₂O₂ production and resulting in a lighter color [61].
- Signal Acquisition: Place the paper device in a standardized imaging box to control lighting. Use the smartphone's camera to capture an image of the detection zone.
- Data Analysis: The dedicated smartphone application processes the image, converting the color intensity into a quantitative value. The concentration of the target pesticide is determined based on a pre-loaded calibration curve, with results displayed on the screen.
Protocol 2: Fluorescence-based Detection Using Solid-State Emitters
This protocol utilizes emissive small molecules for direct visual detection of pesticides on surfaces under a UV lamp.
Workflow Diagram: Solid-State Detection
Materials:
- Stock solution of a solid-state emitter (e.g., TPA-based AIEEgen TT1) in 1,4-dioxane [62].
- Cotton swabs or filter paper strips.
- Portable UV lamp (365 nm).
- Smartphone for documentation (optional).
Procedure:
- Probe Loading: Dip a cotton swab or paper strip into the emitter stock solution (e.g., 10 µM) and allow it to dry briefly.
- Surface Sampling: Gently swab the surface of interest (e.g., fruit skin, soil, or tea leaves).
- Detection: Illuminate the swab or strip with the UV lamp in a darkened environment.
- Result Interpretation: The presence of specific pesticides like trifluralin (TN) or fenitrothion (FN) will cause significant quenching of the probe's fluorescence (e.g., a change from bright green to dim) due to photoinduced electron transfer (PET) and inner-filter effects [62]. This provides a rapid, binary (yes/no) visual result.
Protocol 3: Molecularly Imprinted Polymer (MIP)-based Smartphone Sensing
This protocol leverages the high selectivity and stability of MIPs for detecting pesticides in complex sample matrices.
Workflow Diagram: MIP-based Sensing
Materials:
- MIP nanoparticles specific to the target pesticide (e.g., imidacloprid) [35].
- Optical transducer (e.g., a glass slide or electrode functionalized with the MIP).
- Washing buffer (e.g., phosphate buffer, pH 7.4).
- Smartphone-integrated reading device or adapter.
Procedure:
- Sensor Preparation: Immobilize the synthesized MIPs onto the surface of the transducer. A non-imprinted polymer (NIP) should be prepared in parallel as a control.
- Sample Binding: Incubate the MIP-functionalized transducer with the processed sample extract for a defined period (e.g., 10-15 minutes). The target pesticide molecules will be selectively captured within the complementary cavities of the MIPs.
- Washing: Gently rinse the transducer with buffer to remove any non-specifically bound components from the complex sample matrix.
- Signal Generation and Readout: The binding event is transduced into a measurable signal. This can be a direct color change of the MIP matrix, a fluorescence change of an incorporated dye, or an electrochemical signal. The signal is then captured by the smartphone (either directly or via a simple adapter) and quantified by a dedicated application [35].
Critical Workflow Optimization Strategies
Diagram: Optimization Strategy Logic
To enhance user-friendliness and minimize error, consider these strategies integrated into the protocols:
- Streamline Sample Preparation: Simplify extraction procedures to fewer steps and use solvents compatible with the biosensor elements. For tea matrices, which are complex, this is crucial to avoid interference from polyphenols and alkaloids [1].
- Incorporate Internal Controls: Assay designs should include built-in positive and negative controls. This allows users to verify that the test has functioned correctly, increasing confidence in the result.
- Standardize Readout with Accessories: The use of a simple, 3D-printed dark box or adapter that holds the smartphone at a fixed distance and angle from the sensor ensures uniform lighting and focus, drastically improving the reproducibility of image-based analysis [61].
- Automate Data Interpretation: Rely on smartphone applications to perform all calculations. The user's role should be limited to sample application and image capture, removing subjectivity and potential calculation errors [60] [35]. Future integrations with AI can further enhance this by automatically flagging anomalous results [35].
Application Note
This document provides detailed application notes and experimental protocols for developing smartphone-integrated biosensors for visual pesticide detection. It addresses the critical translational challenges in moving this technology from laboratory proof-of-concept to commercially viable and clinically adopted diagnostic tools, with a specific focus on overcoming manufacturing and regulatory hurdles.
Smartphone-integrated biosensors represent a transformative approach to pesticide detection, offering the potential for rapid, on-site analysis in agricultural, environmental, and food safety contexts. However, a significant gap persists between innovative laboratory prototypes and their widespread clinical and commercial adoption. This gap is characterized by challenges in manufacturing scalability, regulatory approval, and demonstration of clinical utility in real-world settings [5] [63]. The strategies outlined herein are designed to bridge this gap by providing a structured pathway from research to deployment.
Key Biosensing Platforms and Performance Metrics
The selection of an appropriate biosensing platform is fundamental to the success of the application. The table below summarizes the primary biosensing modalities used in conjunction with smartphones for pesticide detection, along with their key performance characteristics.
Table 1: Comparison of Smartphone-Integrated Biosensing Platforms for Pesticide Detection
| Biosensing Platform | Detection Principle | Key Advantages | Reported Limits of Detection (LOD) | Suitable for Scalable Manufacturing? |
|---|---|---|---|---|
| Colorimetric | Measures color change from chemical/biological reaction | Simple, low-cost, uses smartphone camera directly | Varies by assay; can reach picomolar range [32] | High (e.g., paper-based sensors) [64] |
| Electrochemical | Measures electrical signal (current, voltage) from biorecognition event | High sensitivity, miniaturization potential | Not specified in results, but generally high sensitivity [5] | Medium (requires electrode fabrication) |
| Fluorescence | Measures emission light from excited fluorophores | Very high sensitivity, low background | Picomolar range with MOF-enhanced sensors [5] | Medium (requires light source and filters) |
| Bioluminescence | Measures light from enzymatic reaction (e.g., A. fischeri) | No external light source required, low noise | 0.23 ppb for microcystin-LR [64] | High (can be immobilized on paper) [64] |
| Molecularly Imprinted Polymer (MIP) | Synthetic polymers with tailor-made recognition sites | High stability, resistant to harsh conditions | Varies by polymer design and target [35] | High (robust and inexpensive to produce) [35] |
Detailed Experimental Protocols
Protocol 3.1: Fabrication of a Paper-Based Bioluminescent Biosensor
This protocol details the creation of a sustainable, all-in-one paper sensor for toxicity assessment, integrating the bioluminescent bacterium Aliivibrio fischeri [64].
1. Materials and Reagents
- Whatman 1 CHR Chromatography Paper: Serves as the hydrophilic, porous support.
- Wax Printer (e.g., Xerox Phaser 8400): For creating hydrophobic barriers to define sensor wells.
- A. fischeri Bacteria: Bioluminescent bioreporter (available from culture collections).
- Lysogeny Broth (LB) Medium with 3% NaCl: For bacterial culture.
- Agarose: For forming a hydrogel matrix to immobilize bacteria.
- Trehalose and Glycerol: Optional supplements to enhance bacterial stability during storage.
- Adhesive Tape: For sealing the back of the sensor to prevent leakage.
- Cardboard Dark Box (8.5 × 11.5 × 10.0 cm): For blocking ambient light during signal acquisition.
2. Step-by-Step Procedure
- Step 1: Sensor Design and Wax Printing
- Design a circular array of hydrophilic wells (e.g., 3x6 pattern, 7 mm diameter) using presentation software (e.g., PowerPoint).
- Print the design onto the chromatography paper using the wax printer.
- Heat the printed paper at 150°C for 1 minute to allow the wax to penetrate the paper thickness, forming defined hydrophobic boundaries.
Step 2: Bacterial Immobilization
- Culture A. fischeri in LB medium with high salinity at 19°C with orbital shaking (140 rpm) until an OD600 of ~5.0 is reached.
- Prepare a 3% (w/v) agarose solution in sterile water by heating.
- Cool the agarose to approximately 60°C.
- Mix 80 μL of molten agarose with 420 μL of the bacterial suspension (final agarose concentration 0.5% w/v). The final temperature should be ~30°C to avoid harming the bacteria.
- Immediately pipette 20 μL of the bacteria-agarose mixture into each hydrophilic well.
- Allow the hydrogel to solidify by equilibrating at room temperature (25°C) for 30 minutes.
Step 3: Assay Execution and Data Acquisition
- Dispense 30 μL of standard solution or unknown sample into the designated wells.
- Incubate for 1-15 minutes at room temperature.
- Place the sensor inside the dark box to eliminate external light interference.
- Capture the bioluminescent signal using a smartphone camera (e.g., OnePlus 6T) with settings at 30-second integration time and ISO1600.
3. Data Analysis and AI Integration
- Image Processing: Develop a smartphone application (e.g., using Python Kivy framework) to analyze the captured image [64].
- Region of Interest (ROI) Selection: The application automatically identifies and separates the wells containing the colorimetric/bioluminescent reaction using edge detection and color threshold processing.
- Illumination Correction: Apply algorithms like the Gray World algorithm to correct for variations in lighting conditions.
- Quantification: The app interpolates the signal intensity from the sample against an on-board calibration curve to provide a quantitative result (e.g., in toxicity equivalents).
Protocol 3.2: Smartphone-Based Colorimetric Detection with Machine Learning
This protocol leverages smartphone colorimetry and machine learning for precise quantification of pesticide concentrations, overcoming the subjectivity of visual inspection [6].
1. Materials and Reagents
- Gold Nanoparticles (AuNPs): Signal amplification agents in colorimetric reactions.
- Acetylcholinesterase (AChE) Enzyme: Biological recognition element for organophosphate and carbamate pesticides.
- Test Strips: Custom-designed strips with immobilized AChE-AuNPs conjugates.
- Smartphone with Flutter-based Application: For cross-platform deployment.
2. Step-by-Step Procedure
- Step 1: Image Acquisition
- Launch the custom-built Flutter application.
- Use the smartphone's camera API to capture an image of the reacted test strip. The interface should include an overlay to guide alignment.
- Ensure stable exposure and lighting conditions, or use the app's built-in correction.
Step 2: Image Preprocessing
- The application automatically crops the image to the region of interest using color segmentation and contour detection.
- Perform illumination correction using either the Gray World algorithm or a color card calibration method to generate a 3x3 color conversion matrix for accurate color mapping.
Step 3: Feature Extraction and Machine Learning Prediction
- Extract normalized color feature vectors (e.g., RGB, HSV values) from the processed image.
- Feed the feature vector into a pre-trained Support Vector Machine (SVM) model (deployed as an ONNX file) for classification into discrete pesticide concentration levels.
- Refine the quantitative result using a Support Vector Regression (SVR) model, which maps the color features to a continuous concentration value. The final output is a weighted combination of the SVM and SVR results to ensure accuracy and prevent over-prediction.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents and Materials for Biosensor Development
| Item | Function/Application | Key Characteristics |
|---|---|---|
| Molecularly Imprinted Polymers (MIPs) | Synthetic recognition element for pesticides [35] | High selectivity, stability, resistant to denaturation. |
| Gold Nanoparticles (AuNPs) | Colorimetric signal transduction and amplification [5] [6] | High signal amplification efficiency, can be functionalized. |
| Lipid Nanoparticles (LNPs) | Encapsulation and delivery of sensitive bioreporters or enzymes [63] | Protects biological elements, enhances stability. |
| Aliivibrio fischeri | Bioluminescent bioreporter for general toxicity screening [64] | Broad sensitivity to toxins, reproducible response. |
| Acetylcholinesterase (AChE) | Enzyme-based recognition for organophosphates/carbamates [6] | High specificity to a major class of pesticides. |
| CRISPR/Cas12a System | Ultra-sensitive nucleic acid detection for specific toxins [5] | Extremely high sensitivity (fg levels), high specificity. |
| Poly(lactic-co-glycolic acid) (PLGA) | Biopolymer for controlled-release formulations [63] | Biodegradable, biocompatible, tunable release kinetics. |
Strategies for Scalable Manufacturing and Clinical Adoption
Overcoming Manufacturing Hurdles
- Adopt Planar Fabrication Techniques: Utilize wax printing and inkjet printing to create paper-based sensors. These methods are highly scalable, cost-effective, and allow for the mass production of disposable sensor strips [64].
- Embrace Design-for-Manufacturing (DfM): Simplify sensor designs to minimize the number of components and assembly steps. For example, integrating the calibration curve and sample well onto a single paper substrate streamlines production and use [64].
- Ensure Batch-to-Batch Consistency: Implement rigorous quality control measures for critical sensor components. The use of nanomaterials like gold nanoparticles has demonstrated low inter-batch coefficients of variation (below 5%), which is essential for reproducibility [5].
- Develop Stable Bioreceptor Formulations: Invest in advanced formulation strategies to enhance the shelf-life of biological elements. This includes immobilizing bacteria in hydrogel matrices [64] and exploring non-PEG alternatives for surface functionalization to avoid immunogenic responses [63].
Navigating the Clinical and Regulatory Pathway
- Demonstrate Analytical and Clinical Validity: Conduct rigorous testing with real-world samples (e.g., spiked tap water, wastewater, crop extracts) to validate the sensor's performance against gold-standard methods like GC-MS or HPLC-MS/MS [1] [64]. The biosensor must demonstrate high sensitivity, specificity, and a low false-positive rate.
- Address "One-Sensor-Fits-All" Variability: Acknowledge that performance can vary across different smartphone models. Develop robust software that uses internal calibration and AI-driven correction algorithms to normalize results across various camera resolutions and lighting conditions [6] [64].
- Plan for Regulatory Approval Early: Engage with regulatory bodies (FDA, EMA) during the development phase. Prepare a detailed dossier covering Chemistry, Manufacturing, and Controls (CMC), along with data from biocompatibility and toxicity studies, as required for complex non-biological products [63].
- Focus on User-Centered Design: Create intuitive smartphone applications with simple user interfaces, large buttons, and clear instructions to ensure usability for non-experts in field conditions [6]. This is critical for patient adoption and compliance.
Visual Workflows for Biosensor Development and Deployment
The following diagrams illustrate the core workflows for biosensor operation and the critical path from development to clinical adoption.
Diagram 1: Biosensor operational workflow from sample to result.
Diagram 2: Strategic pathway for translational development.
Benchmarking Performance: Validation, Regulatory Hurdles, and Future Commercial Viability
In the development and validation of smartphone-integrated biosensors for visual pesticide detection, three analytical performance metrics are paramount: the Limit of Detection (LOD), Sensitivity, and Specificity. These parameters mathematically describe the accuracy, reliability, and practical utility of a biosensing system. For researchers aiming to deploy these biosensors in field settings, a deep understanding of the interrelationship and trade-offs between these metrics is critical. The LOD defines the lowest concentration of an analyte that the biosensor can reliably detect, while sensitivity reflects the test's ability to correctly identify true positive samples (e.g., pesticide-contaminated samples), and specificity indicates its ability to correctly identify true negative samples (e.g., pesticide-free samples) [65] [66]. Within the framework of a smartphone-based platform, these metrics are influenced by factors including biorecognition element affinity, transducer signal-to-noise ratio, and the performance of the mobile algorithm for data interpretation [5]. This document provides detailed application notes and protocols for determining and optimizing these metrics, specifically tailored for visual pesticide detection research.
Conceptual Foundations and Definitions
Limit of Detection (LOD)
The Limit of Detection is the lowest concentration of a pesticide that can be consistently distinguished from a blank sample (containing no pesticide). It is a measure of the ultimate detectability of the assay.
- Mathematical Definition: Typically, LOD is calculated as three times the standard deviation of the blank (or negative control) signal divided by the slope of the calibration curve: LOD = 3σ/S, where σ is the standard deviation of the blank measurement, and S is the slope of the calibration curve.
- Context in Visual Pesticide Detection: For smartphone-based colorimetric sensors, the "signal" is often a change in color intensity, channel value (e.g., RGB, HSV), or a derived pixel intensity value. A key consideration is the LOD paradox: achieving an ultra-low LOD is a celebrated technical feat, but it must align with the clinically or environmentally significant concentration range. For many pesticides, detection well below the maximum residue limit (MRL) is essential, but an excessively low LOD may complicate the assay without adding practical value [66].
Sensitivity
Sensitivity, also known as the true positive rate, measures the proportion of actual positive samples that are correctly identified as positive by the biosensor [65].
- Mathematical Definition: Sensitivity is calculated as the number of true positives divided by the sum of true positives and false negatives.
Sensitivity = Number of True Positives / (Number of True Positives + Number of False Negatives) - Implication: A test with 100% sensitivity means it correctly identifies every contaminated sample. In a high-sensitivity test, a negative result is powerful for "ruling out" the presence of a pesticide above the LOD [65].
Specificity
Specificity, or the true negative rate, measures the proportion of actual negative samples that are correctly identified as negative [65].
- Mathematical Definition: Specificity is calculated as the number of true negatives divided by the sum of true negatives and false positives.
Specificity = Number of True Negatives / (Number of True Negatives + Number of False Positives) - Implication: A test with 100% specificity means no pesticide-free samples are incorrectly flagged as contaminated. A positive result in a high-specificity test is reliable for "ruling in" the presence of the pesticide [65].
Trade-offs and Relationships
There is an inherent trade-off between sensitivity and specificity, often governed by the chosen detection threshold. Setting a very low threshold for a positive signal may increase sensitivity but also increases false positives, thereby reducing specificity. Conversely, a high threshold can maximize specificity at the cost of missing some true positives (lower sensitivity) [65]. The LOD establishes the lower boundary of the dynamic range within which these trade-offs are negotiated. A focus on pushing the LOD to ultra-low levels can sometimes come at the expense of a robust dynamic range or the assay's simplicity and cost-effectiveness [66].
Table 1: Summary of Core Analytical Performance Metrics
| Metric | Definition | Key Question Answered | Ideal Value |
|---|---|---|---|
| Limit of Detection (LOD) | The lowest analyte concentration reliably distinguished from a blank. | How low can you detect? | As low as the regulatory limit requires. |
| Sensitivity | The ability to correctly identify positive samples. | How well do you find the contaminant? | 100% (1.0) |
| Specificity | The ability to correctly identify negative samples. | How well do you avoid false alarms? | 100% (1.0) |
Experimental Protocols for Metric Determination
Protocol for Determining Limit of Detection (LOD)
This protocol outlines the procedure for determining the LOD of a smartphone-integrated colorimetric biosensor for a target pesticide.
1. Principle The LOD is estimated by analyzing the response of both blank samples (negative controls) and low-concentration calibration standards. The signal is measured as a color intensity value derived from the smartphone's camera.
2. Research Reagent Solutions & Materials Table 2: Key Reagents and Materials for LOD Determination
| Item | Function/Description |
|---|---|
| Pesticide Standard | High-purity analytical standard of the target pesticide for preparing calibration solutions. |
| Sample Matrix | The background solution in which the pesticide is dissolved (e.g., buffer, purified water, or a representative food extract). |
| Colorimetric Probe | The reagent that produces a color change upon interaction with the pesticide (e.g., enzyme, antibody, molecularly imprinted polymer with chromogenic substrate). |
| Smartphone with App | A mobile device with a dedicated application for image capture, color analysis (RGB extraction), and data processing. |
| Static Imaging Box | A light-controlled chamber to ensure consistent, reproducible imaging conditions and minimize ambient light interference. |
3. Procedure
- Step 1: Preparation of Calibration Standards. Prepare a series of pesticide standard solutions in the relevant sample matrix, spanning a concentration range from expected zero to a level above the anticipated LOD. Include at least five concentration levels and a minimum of six replicate blank samples (containing all reagents except the pesticide).
- Step 2: Assay Execution. For each calibration standard and blank, perform the complete colorimetric assay as per the developed protocol (e.g., mix probe, incubate, etc.).
- Step 3: Image Acquisition and Signal Processing. Place each reaction solution in the static imaging box. Use the smartphone app to capture an image under standardized lighting and camera settings. The app should extract a predefined color signal (e.g., the Red value, Blue value, or a calculated grayscale intensity) from a defined region of interest for each sample.
- Step 4: Data Calculation.
- Calculate the mean signal (
Mean_blank) and standard deviation (SD_blank) of the six blank replicates. - Generate a calibration curve by plotting the signal (y-axis) against the pesticide concentration (x-axis). Perform linear regression to obtain the slope (
S) of the linear portion of the curve. - Calculate the LOD using the formula: LOD = (3.3 × SD_blank) / S. The factor 3.3 provides a 99% confidence level.
- Calculate the mean signal (
4. Data Analysis The calculated LOD should be verified experimentally by analyzing several samples spiked with pesticide at the LOD concentration. The observed signal for these samples should be statistically distinguishable from the blank signal.
Protocol for Determining Sensitivity and Specificity
This protocol uses a validation set of samples with known pesticide status to characterize the biosensor's diagnostic performance.
1. Principle Sensitivity and specificity are calculated by comparing the biosensor's results against a reference method (the "gold standard") for a set of samples with known concentrations.
2. Research Reagent Solutions & Materials The materials listed in Table 2 are also required. Additionally, a set of characterized samples (e.g., spiked and unspiked samples validated by a reference method like GC-MS) is essential.
3. Procedure
- Step 1: Assay Validation Set. Prepare or obtain a panel of samples. The panel should include a representative number of samples known to be positive (contaminated with pesticide at or above the LOD) and negative (pesticide-free or below the LOD), as confirmed by a gold-standard method.
- Step 2: Blind Testing. Analyze each sample in the validation set using the smartphone-integrated biosensor protocol without knowledge of the known concentration (blind analysis).
- Step 3: Result Classification. For each sample, classify the biosensor's result as positive or negative based on a pre-defined signal threshold. Compare this result to the sample's known status to populate a confusion matrix (True Positive, False Positive, True Negative, False Negative).
4. Data Analysis
- Calculate Sensitivity = TP / (TP + FN)
- Calculate Specificity = TN / (TN + FP)
- The 95% confidence intervals for these proportions should be calculated to understand the precision of the estimates.
Visualization of Metrics and Workflow
The following diagrams, created using Graphviz, illustrate the core concepts and experimental workflow.
Relationship between LOD, Sensitivity, and Specificity
Experimental Workflow for Metric Determination
Application in Smartphone-Integrated Biosensors
The integration of biosensors with smartphones introduces specific considerations for these performance metrics. The smartphone serves as a power source, processor, display, and communication hub, but its variability can impact performance [5]. Key challenges include:
- LOD and Signal Amplification: Achieving a low LOD often requires signal amplification strategies compatible with smartphone detection. These include using nanomaterials like gold nanoparticles (which can boost signal efficiency by up to 50% [5]) or enzymatic amplification. The smartphone's camera must be sensitive enough to detect the resulting colorimetric changes. The use of microfluidic chips can automate sample handling and improve reproducibility, aiding in precise LOD determination [5].
- Sensitivity/Specificity and Assay Chemistry: The fundamental sensitivity and specificity of the system are primarily determined by the biorecognition element (e.g., the affinity of an antibody or the specificity of an enzyme for the target pesticide). However, the smartphone's role in data analysis is crucial. AI/ML-powered mobile platforms can enhance diagnostic robustness by analyzing complex outputs and reducing misinterpretation, potentially improving effective sensitivity and specificity [5].
- Calibration and Standardization: A significant barrier to deployment is the lack of standardized calibration protocols across different smartphone models, which can lead to performance variability [5]. For sensitivity and specificity to hold true across devices, consistent color measurement and processing are essential. This often necessitates the use of a static imaging box to control lighting and a reference color chart within each image for normalization.
In conclusion, the rigorous characterization of LOD, Sensitivity, and Specificity is non-negotiable for developing a credible smartphone-integrated biosensor for pesticide detection. The protocols and frameworks provided here offer a pathway to achieving this, ensuring that the technology is not only technically sound but also fit for its intended purpose in real-world applications [66].
The increasing demand for rapid, on-site detection of environmental contaminants, particularly pesticides, has accelerated the development of smartphone-integrated biosensors. However, validating these novel platforms requires rigorous comparison against established analytical techniques. This Application Note provides a detailed comparative analysis of three gold-standard methods—High-Performance Liquid Chromatography (HPLC), Gas Chromatography-Mass Spectrometry (GC-MS), and Enzyme-Linked Immunosorbent Assay (ELISA)—within the context of validating smartphone-based biosensors for pesticide detection. We present standardized protocols and performance data to guide researchers in selecting appropriate reference methods for biosensor validation, ensuring data reliability and regulatory compliance.
Comparative Performance of Analytical Techniques
The selection of an appropriate reference method is critical for the validation of novel biosensing platforms. Table 1 summarizes the key analytical performance parameters of HPLC, GC-MS, and ELISA methods as documented in recent literature for pesticide detection.
Table 1: Performance Comparison of Gold-Standard Methods for Pesticide Analysis
| Method | Typical Analytes | Detection Limit | Recovery Range | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| HPLC-MS/MS | Multi-class pesticides (carbamates, organophosphates, organochlorines, pyrethroids) [67] | ~0.0005 mg/kg [68] | 70-119% [67] | High sensitivity, broad multi-residue capability, structural confirmation | High instrument cost, requires skilled operators, extensive sample preparation |
| GC-MS/MS | GC-amenable pesticides (volatile, non-polar) [68] | 0.001 mg/kg [68] | Most analytes within SANTE recovery tolerances [68] | Excellent sensitivity for volatile compounds, robust compound libraries | Limited to volatile/thermostable compounds, derivatization sometimes needed |
| ELISA | Class-specific pesticides (e.g., organophosphates [69], imidacloprid [70]) | Varies by analyte (e.g., 6.3 ng/mL for parathion [69]) | 75.7-105.3% [69] | High throughput, lower cost, minimal sample cleanup | Potential cross-reactivity, higher variability vs. chromatographic methods [71] [72] |
| Smartphone Biosensor | Organophosphates (e.g., Paraoxon-Methyl) [10] | 0.304 ppt (Paraoxon-Methyl) [10] | 98.3% average recovery [10] | Portability, rapid on-site analysis, user-friendly operation | Limited multiplexing, developing technology requiring validation |
Independent comparative studies highlight important considerations for method selection. When comparing LC-MS/MS with ELISA for biomarker analysis, LC-MS/MS demonstrated superior accuracy with measurements 7.6- to 23.5-fold lower than ELISA results, though correlation improved with solid-phase extraction (SPE) purification [72]. Similarly, for cotinine detection, LC-MS/MS showed enhanced sensitivity (LOQ: 0.1 ng/mL) compared to ELISA (LOQ: 0.15 ng/mL) and revealed associations with demographic variables that ELISA failed to detect [71].
Experimental Protocols
HPLC-MS/MS Method for Pesticide Residues in Food Matrices
This protocol follows SANTE/11312/2021v2 guidance for determining 121 pesticide residues in rice, adapted from the study by da Silva et al. [67].
Reagents and Solutions
- Pesticide standards: Certified reference materials for target analytes
- Extraction solvent: Acetonitrile, HPLC grade
- QuEChERS salts: 4 g MgSO₄, 1 g NaCl, 0.5 g disodium citrate sesquihydrate, 1 g sodium citrate tribasic dihydrate per sample
- dSPE clean-up: 150 mg MgSO₄, 25 mg PSA per 1 mL extract
- Mobile phases: A) 5 mM ammonium formate in water, B) 5 mM ammonium formate in methanol
Sample Preparation (QuEChERS)
- Homogenization: Homogenize 10 g representative sample with 10 mL acetonitrile in a 50 mL centrifuge tube.
- Extraction: Add QuEChERS salt mixture, shake vigorously for 1 minute, then centrifuge at 4000 rpm for 5 minutes.
- Clean-up: Transfer 1 mL supernatant to dSPE tube containing MgSO₄ and PSA. Shake for 30 seconds and centrifuge at 4000 rpm for 5 minutes.
- Preparation for analysis: Transfer 0.5 mL cleaned extract to autosampler vial, dilute with 0.5 mL water if necessary.
Instrumental Analysis
- HPLC system: Agilent 1200 series or equivalent
- Column: C18 reversed-phase (e.g., 100 mm × 2.1 mm, 1.8 μm)
- Mass spectrometer: Triple quadrupole MS with ESI source (e.g., API 3200)
- Gradient program:
- 0-1 min: 5% B
- 1-10 min: 5-95% B
- 10-13 min: 95% B
- 13-13.1 min: 95-5% B
- 13.1-16 min: 5% B (re-equilibration)
- Flow rate: 0.3 mL/min
- Injection volume: 5 μL
- Ionization mode: ESI positive/negative switching
- Detection: Multiple Reaction Monitoring (MRM) with compound-specific transitions
Method Validation
- Linearity: Prepare matrix-matched calibration standards (0.0005-0.1 mg/kg). Acceptable regression coefficient (r²) > 0.99.
- Recovery: Fortify blank matrix at 0.01 mg/kg and 0.1 mg/kg (n=5). Calculate recovery as (measured concentration/spiked concentration) × 100%. Acceptable range: 70-120% [67].
- Precision: Calculate relative standard deviation (RSD) of recovery samples. Acceptable RSD ≤ 20%.
GC-MS/MS Method for Pesticide Residues in Cucumber
This protocol, adapted from Waters Application Note [68], enables detection of over 200 GC-amenable pesticides.
Sample Preparation
- Extraction: Weigh 10 g homogenized cucumber into 50 mL centrifuge tube. Add 10 mL acetonitrile and QuEChERS salts.
- Shaking and centrifugation: Shake vigorously for 1 minute, then centrifuge at 4000 rpm for 5 minutes.
- dSPE clean-up: Transfer 1 mL upper layer to dSPE tube (150 mg MgSO₄, 25 mg PSA). Shake for 30 seconds and centrifuge.
GC-MS/MS Analysis
- GC system: Agilent 7890A or equivalent
- Injector: Pulsed splitless mode, 250°C, 1 μL injection
- Column: Rxi-5Sil MS (30 m × 0.25 mm id × 0.25 μm)
- Carrier gas: Helium, constant flow (2 mL/min)
- Oven program:
- 90°C for 1 min
- 90°C to 330°C at 8.5°C/min
- 330°C for 5 min
- Mass spectrometer: Xevo TQ-XS with APGC source
- Source temperature: 150°C
- Corona current: 2.0 μA
- Detection: MRM mode with compound-specific transitions
ELISA for Organophosphorus Pesticides in Camellia Oil
This protocol describes a matrix solid-phase dispersion and direct competitive ELISA for five organophosphorus pesticides in camellia oil [69].
Immunoassay Procedure
- Coating: Coat microplate wells with 100 μL/well of appropriate capture antibody in coating buffer. Incubate overnight at 4°C.
- Blocking: Wash plates 3 times with PBST, then add 200 μL/well blocking buffer (1% BSA in PBS). Incubate 1 hour at 37°C.
- Competition: Add 50 μL/well of standard or sample extract plus 50 μL/well of enzyme conjugate (hapten-HRP). Incubate 30 minutes at 37°C.
- Detection: Wash plates 5 times with PBST. Add 100 μL/well substrate solution (TMB/H₂O₂). Incubate 15-30 minutes at room temperature.
- Stopping and measurement: Add 50 μL/well stop solution (2M H₂SO₄). Measure absorbance at 450 nm.
Matrix Solid-Phase Dispersion (MSPD)
- Column preparation: Place C18 sorbent in SPE cartridge.
- Sample preparation: Blend 0.5 g oil sample with 0.5 g C18 sorbent until homogeneous.
- Transfer and elution: Transfer mixture to SPE cartridge. Elute target pesticides with 10 mL acetonitrile.
- Concentration: Evaporate eluate to dryness under nitrogen, reconstitute in assay buffer for ELISA.
Integration with Smartphone Biosensor Development
The validation of emerging smartphone-integrated biosensors requires careful comparison with these established methods. Figure 1 illustrates the typical validation workflow for a smartphone biosensor against gold-standard methods.
Figure 1: Workflow for validating smartphone biosensors against gold-standard methods.
Recent advances in smartphone-integrated electrochemical biosensors demonstrate the potential of these platforms. One study reported an acetylcholinesterase-based biosensor coupled with a mobile app that achieved a detection limit of 0.304 ppt for Paraoxon-Methyl with an average recovery of 98.3% in food/water samples, showing comparable results to LC-MS/MS [10]. These biosensors leverage the hydrolytic activity of acetylcholinesterase, where pesticide inhibition reduces proton doping of polyaniline nanofibers, decreasing conductance measurable by the device [10].
Table 2 outlines essential research reagents and materials critical for implementing these analytical methods in biosensor validation studies.
Table 2: Research Reagent Solutions for Analytical Methods
| Reagent/Material | Function | Example Specifications | Application Areas |
|---|---|---|---|
| QuEChERS Extraction Kits | Multi-residue pesticide extraction | Contains MgSO₄, NaCl, citrate salts, PSA [67] | Sample preparation for HPLC-MS/MS, GC-MS/MS |
| C18 Chromatography Columns | Reversed-phase separation | 100 mm × 2.1 mm, 1.8 μm particle size [67] | HPLC separation of pesticide residues |
| APGC Source | Soft ionization for GC-MS | Atmospheric pressure chemical ionization [68] | Improved sensitivity for GC-amenable pesticides |
| Polyclonal Antibodies | Molecular recognition | Specific to pesticide haptens (e.g., imidacloprid) [70] | ELISA development, biosensor recognition elements |
| Acetylcholinesterase Enzyme | Biosensor recognition element | Inhibited by organophosphate pesticides [10] | Smartphone biosensors for OPs |
| Polyaniline Nanofibers (PAnNFs) | Signal transduction | Conductance changes with proton doping [10] | Resistive biosensors for OPs |
| Gold Interdigitated Electrodes | Biosensor platform platform | Microfabricated electrode patterns [10] | Electrochemical biosensor development |
This Application Note provides comprehensive protocols for gold-standard analytical methods relevant to the validation of smartphone-integrated biosensors for pesticide detection. The comparative data demonstrates that while HPLC-MS/MS offers the highest sensitivity and broadest analyte coverage, ELISA provides a cost-effective alternative for high-throughput screening, and emerging smartphone biosensors show promising potential for rapid on-site detection. The choice of reference method should be guided by the specific validation requirements, target analytes, and intended application of the biosensor platform. As smartphone-based detection technologies continue to evolve, rigorous validation against these established methods will be essential for regulatory acceptance and field deployment.
Validation with Real-World Samples and Reproducibility Across Labs
The following tables consolidate key quantitative data from recent studies on smartphone-integrated biosensors for pesticide detection, focusing on analytical performance and validation results.
Table 1: Analytical Performance of Smartphone-Integrated Biosensors for Pesticide Detection
| Biosensor Platform | Target Analyte | Sample Matrix | Linear Range | Limit of Detection (LOD) | Reproducibility (RSD) | Reference |
|---|---|---|---|---|---|---|
| Smartphone/Resistive Nanosensor (AChE/MWCNT-PAnNF) | Paraoxon-Methyl | Food, Water | 1 ppt – 100 ppb | 0.304 ppt | < 5% | [10] |
| Smartphone/Resistive Nanosensor (AChE/BChE/MWCNT-PAnNF) | Organophosphate Pesticides (via enzyme inhibition) | Finger-stick Blood | AChE: 2.0–18.0 U/mLBChE: 0.5–5.0 U/mL | AChE: 0.11 U/mLBChE: 0.093 U/mL | < 4% | [23] |
| CRISPR/Cas12a-based Platform | Specific DNA Targets | - | - | 40 femtograms/reaction | - | [5] |
| Gold Nanoparticle-enhanced Electrochemical Biosensor | - | - | - | - | < 5% (inter-batch CV) | [5] |
Table 2: Validation with Real-World Samples: Recovery and Comparative Analysis
| Biosensor Platform | Target Analyte | Real-World Sample Type | Average Recovery Rate | Validation Method | Key Finding | Reference |
|---|---|---|---|---|---|---|
| Smartphone/Resistive Nanosensor | Paraoxon-Methyl | Food, Environmental Water | 98.3% | Liquid Chromatography-Mass Spectrometry (LC-MS) | Strong agreement with standard method | [10] |
| Smartphone/Resistive Nanosensor (AChE/BChE) | Organophosphate Pesticides (Exposure) | Farmworker Blood (n=22) | - | Radiometric Method, Ellman's Method | Strong agreement with both standard methods | [23] |
Experimental Protocols
Protocol: Fabrication of CS/MWCNT/PAnNF-Modified Gold Interdigitated Electrode (AuIDE) Nanosensor
This protocol details the synthesis of the core sensing element used in resistive biosensors for organophosphate detection [23].
Materials:
- Chitosan (CS)
- Multi-walled carbon nanotubes (MWCNTs)
- Polyaniline nanofibers (PAnNFs)
- Gold Interdigitated Electrodes (AuIDEs)
- Glass fiber (GF) pads (Ø 2.5 mm and Ø 4/3 mm)
- Anti-interference reagents: BW284c51 (AChE-specific inhibitor), MgCl₂, CaCl₂
- Enzyme substrates: Acetylcholine (ACh) or Butyrylcholine (BCh)
Procedure:
- AuIDE Cleaning: Clean the Gold Interdigitated Electrodes (AuIDEs) chemically to ensure a pristine surface for modification.
- Nanocomposite Preparation: Synthesize the MWCNT/PAnNF nanomaterial. Prepare a homogeneous composite by integrating the nanomaterials with chitosan.
- Electrode Modification: Deposit the CS/MWCNT/PAnNF nanocomposite film onto the cleaned surface of the AuIDE to form the transducer.
- Reagent Pad Assembly:
- Inner Pad: Pre-load a Ø 2.5 mm GF pad with the substrate (ACh or BCh).
- Outer Pad: Pre-load a larger GF pad (outer Ø 4 mm, inner Ø 3 mm) with anti-interference reagents (e.g., BW284c51, MgCl₂ + CaCl₂).
- Biosensor Integration: Assemble the modified AuIDE with the outer and inner pre-loaded reagent pads into a single, integrated platform.
Protocol: On-Site Biomontoring of OP Pesticide Exposure in Whole Blood
This protocol describes the application of the integrated nanosensor for rapid detection of pesticide exposure from a drop of blood [23].
Materials:
- Integrated smartphone/resistive nanosensor platform (from Protocol 2.1)
- Bluetooth resistance meter
- Smartphone with dedicated application installed
- Finger-stick whole blood sample
Procedure:
- Sample Introduction: Apply a drop of finger-stick whole blood directly to the biosensor platform. The integrated design requires no external sample preprocessing.
- Reaction Initiation: The blood sample hydrates the pre-loaded reagents in the outer and inner pads. The substrate (ACh/BCh) is hydrolyzed by the native AChE/BChE present in the blood, generating protons.
- Signal Transduction: The generated protons dope the PAnNFs in the nanocomposite film, changing its electrical conductance.
- Signal Measurement: The Bluetooth resistance meter records the change in resistance (conductance) of the nanosensor in real-time.
- Data Processing & Display: The smartphone application processes the resistance data, calculates the cholinesterase enzyme activity, and displays the result within approximately 10 minutes. The app also stores, tracks, and enables sharing of the results.
Visualized Workflows and Signaling Pathways
Biosensor Operation Workflow
This diagram illustrates the end-to-end process from sample application to result output for smartphone-integrated biosensors.
Resistive Signal Generation Logic
This diagram explains the correlation between pesticide concentration, enzyme activity, and the resulting electrical signal that the smartphone measures.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials and Reagents for Smartphone-Integrated Pesticide Biosensors
| Item | Function/Role in the Experiment | Reference |
|---|---|---|
| Acetylcholinesterase (AChE) / Butyrylcholinesterase (BChE) | Primary biological recognition element. OP pesticides inhibit its activity, which is the basis for detection. | [23] [10] |
| Polyaniline Nanofibers (PAnNFs) | Key transducer material. Its electrical conductance increases when doped by protons (H⁺) generated from enzyme-substrate hydrolysis. | [23] [10] |
| Multi-walled Carbon Nanotubes (MWCNTs) | Nanomaterial used in the nanocomposite to enhance electrical conductivity and provide a high-surface-area scaffold. | [23] |
| Chitosan (CS) | A biopolymer used to form a stable matrix for the MWCNT/PAnNF nanocomposite on the electrode surface. | [23] |
| Gold Interdigitated Electrode (AuIDE) | The physical transducer platform that measures the resistance change of the deposited nanocomposite film. | [23] |
| Acetylcholine (ACh) / Butyrylcholine (BCh) | Enzyme substrates. Their hydrolysis by AChE/BChE produces the protons that dope the PAnNFs. | [23] [10] |
| BW284c51 | A specific inhibitor of AChE. Used in reagent pads to create selectivity for BChE measurement in complex samples like blood. | [23] |
The integration of smartphones as analytical platforms for visual pesticide detection represents a paradigm shift in agricultural biosensing. These systems leverage smartphone cameras, processors, and connectivity to enable rapid, on-site quantification of pesticide residues, offering a powerful alternative to traditional laboratory methods [32]. However, the journey from a validated laboratory prototype to a commercially available product is complex, governed by stringent regulatory frameworks and a dynamic intellectual property landscape. For researchers and developers, navigating this path is critical for successful technology transfer and market adoption. This document outlines the essential regulatory considerations, patent trends, and strategic protocols for advancing smartphone-integrated biosensors for visual pesticide detection toward commercialization.
Regulatory Landscape for Commercialization
Navigating the regulatory environment is a fundamental step in the commercialization process. Regulatory bodies ensure that diagnostic devices are safe, effective, and reliable for end-users.
Key Regulatory Considerations
Smartphone-based biosensors for pesticide detection are typically classified as in vitro diagnostic devices. Their regulatory pathway is influenced by their intended use, target analyte, and potential impact on public health. A primary challenge is that these devices often combine hardware (sensors, attachments), software (mobile apps, algorithms), and biological components (enzymes, aptamers), each of which may fall under different regulatory scrutiny [3] [73]. The software component, which handles data acquisition, processing, and result interpretation, must be validated to ensure analytical robustness and cybersecurity, particularly if it integrates with telehealth platforms or electronic health records [5] [74].
A significant barrier is the performance variability under real-world conditions. Environmental factors such as temperature fluctuations, humidity, and variability in biological samples can distort readings, leading to diagnostic inaccuracies that erode user and regulatory confidence [5]. Furthermore, a lack of standardized calibration protocols and inconsistent signal processing across different smartphone models present a major hurdle to clinical validation and regulatory approval [5]. Developers must generate extensive validation data demonstrating that their device performs consistently across all intended smartphone models and environmental conditions.
Compliance and Validation Strategies
Early engagement with regulatory agencies is crucial to define the appropriate classification and required evidence. A comprehensive quality management system (QMS), such as ISO 13485, should be implemented throughout the development lifecycle. Performance validation must adhere to established guidelines from organizations like the AOAC International or the EPA for pesticide detection methods [73]. Key parameters to establish include:
- Accuracy and Precision: Demonstrated through comparison with reference standard methods (e.g., GC-MS, LC-MS) using spiked and real-world samples.
- Limit of Detection (LOD) and Quantification (LOQ): Must be sufficiently low to meet regulatory thresholds for pesticide residues in food.
- Robustness and Stability: Testing should account for different lighting conditions, camera specifications, and sample matrix effects [32] [43].
- Software Verification and Validation: The mobile application must be rigorously tested for accurate data processing, secure data handling, and user interface reliability [73].
Patent Trends and Intellectual Property Strategy
The intellectual property landscape for smartphone-based biosensors is dynamic, reflecting both technological innovation and market realities.
Analysis of Current Patent Trends
Historically, patent filings for smartphone-based biosensors saw significant growth until approximately 2016, followed by a notable decline in subsequent years [3]. This trend may be attributed to the "Theranos effect," which increased investor and regulatory skepticism towards novel diagnostic platforms, and the technical difficulty of transitioning laboratory validations to commercially viable products with real-world samples [3].
Globally, the United States leads in patent filings, with over 3,000 patents focused on advanced sensor technologies, wireless communication, and AI integration [74]. Europe shows strong activity in data security and device interoperability, driven by stringent regulations, while the Asia-Pacific region emphasizes cost-effective and scalable solutions [74]. Leading corporate players, such as Medtronic and Koninklijke Philips, hold thousands of patents, focusing on wearable technology, AI-driven monitoring, and telehealth integration [74]. This indicates a highly competitive and mature landscape in adjacent diagnostic fields, which new entrants must navigate carefully.
Strategic IP Management for Researchers
For research teams, a proactive IP strategy is essential:
- Conduct Thorough Prior Art Searches: Before filing, perform comprehensive searches to ensure novelty and avoid infringement. The report notes that over 6,000 patents have been filed globally in related digital monitoring fields [74].
- File Early and Broadly: Given the competitive landscape, file provisional patents early to secure priority dates. Consider international protection in key markets like the U.S., Europe, and Asia.
- Focus on Technical Differentiators: Key patentable areas include novel biorecognition elements (e.g., specific aptamers for pesticides), unique microfluidic designs for sample preparation, proprietary signal amplification strategies using nanomaterials, and specialized data analytics algorithms for image processing and concentration quantification [5] [43].
- Consider Collaboration: Partnerships with established entities can provide access to broader IP portfolios and manufacturing expertise.
Table 1: Key Patent Trends in Smartphone-Based Diagnostic Devices
| Aspect | Trend and Observation | Strategic Implication |
|---|---|---|
| Overall Filing Volume | Rapid growth until ~2016, followed by a significant decline [3]. | Highlights market challenges; underscores need for robust, commercially viable inventions. |
| Geographical Distribution | US leads (>3,000 patents), followed by Europe and Asia-Pacific [74]. | Requires a global IP strategy, with filings in key technological and commercial markets. |
| Key Technologies | Wireless sensors, AI integration, wearable devices, telehealth solutions [74]. | Innovation should be directed towards integrating these high-value, trending technologies. |
| Dominant Players | Medtronic, Koninklijke Philips, etc., hold large patent portfolios [74]. | New entrants must conduct careful freedom-to-operate analyses and find niche applications. |
Experimental Protocols for Regulatory and Patent Validation
Generating robust experimental data is critical for both regulatory submissions and patent applications to demonstrate utility and non-obviousness.
Protocol: Analytical Performance Validation
This protocol outlines the key experiments required to validate the analytical performance of a smartphone-based biosensor for pesticide detection.
1. Objective: To determine the Limit of Detection (LOD), Limit of Quantification (LOQ), linear dynamic range, accuracy, and precision of the biosensor. 2. Materials:
- Smartphone-based biosensor platform with integrated app for RGB/HSV analysis.
- Microfluidic chips or paper-based substrates (μPADs).
- Biorecognition element (e.g., acetylcholinesterase for organophosphates, specific aptamer).
- Standard solutions of target pesticide (e.g., chlorpyrifos) in a suitable solvent.
- Negative control samples (pesticide-free buffer and matrix extracts).
- Reference analytical method (e.g., LC-MS/MS). 3. Procedure:
- Calibration Curve: Prepare a series of standard pesticide solutions across a concentration range (e.g., 0.1 ppb to 1000 ppb). For each concentration, perform the assay in triplicate. Use the smartphone app to capture images and extract color values (e.g., R, G, B, or Hue). Plot the color intensity (or ratio) versus the logarithm of the concentration to generate a calibration curve.
- LOD and LOQ Calculation: LOD can be calculated as 3.3σ/S and LOQ as 10σ/S, where σ is the standard deviation of the response of the blank and S is the slope of the calibration curve.
- Accuracy and Precision: Spike pesticide-free food samples (e.g., apple extract) with known concentrations of the pesticide at low, medium, and high levels within the dynamic range. Analyze these samples (n=5 per concentration) to determine intra-day precision. Repeat the analysis on three different days to determine inter-day precision. Accuracy is reported as percent recovery of the spiked amount.
- Cross-Reactivity: Test the biosensor against other common pesticides and matrix interferents to establish specificity.
Protocol: Robustness and Real-World Testing
1. Objective: To assess the performance of the biosensor across different smartphone models and under varying environmental conditions. 2. Materials: Multiple smartphone models (varying camera resolution and age), portable temperature and light control chamber. 3. Procedure:
- Inter-smartphone Variability: Use the same calibrated assay and run identical samples on 5-10 different smartphone models. Analyze the results to determine the coefficient of variation (CV) across devices. A CV below 10-15% is typically desirable.
- Environmental Robustness: Perform assays at different temperatures (e.g., 15°C, 25°C, 35°C) and lighting conditions (e.g., dim, fluorescent, bright outdoor). Quantify the impact on the signal output and LOD.
Table 2: Research Reagent Solutions for Biosensor Development
| Reagent/Material | Function in the Experiment | Key Considerations |
|---|---|---|
| Enzymes (e.g., AChE, ChOx) | Biorecognition element; catalytic activity inhibition by pesticides enables detection [43]. | Select for high specificity and stability; immobilization method is critical for activity retention. |
| Aptamers | Synthetic biorecognition element; binds to specific pesticide molecules with high affinity [43]. | Offer advantages over antibodies in stability and production; require SELEX for development. |
| Gold Nanoparticles (AuNPs) | Signal amplification tag; high conductivity and unique optical properties enhance sensitivity [5] [43]. | Control size and functionalization for consistent performance; can boost signal efficiency by up to 50% [5]. |
| Graphene Oxide (GO)/Reduced GO | Nanomaterial for electrode modification; provides large surface area for immobilization and enhances electron transfer [43]. | Oxygen-containing groups enable easy functionalization; reduction state tunes conductivity. |
| Microfluidic Chips (μPADs) | Miniaturized platform for automated fluid handling and sample processing; reduces reagent use [3] [43]. | Design must ensure proper fluidic control and alignment with smartphone optical sensors. |
| Colorimetric Probes | Produces a visual color change upon reaction with the target or a generated product [32]. | Must be selected for a strong, measurable color shift compatible with smartphone camera RGB filters. |
Workflow Visualization
The following diagram illustrates the integrated workflow for development, validation, and commercialization, highlighting the parallel tracks of technical and regulatory activities.
Integrated R&D and Commercialization Workflow
The path to market for smartphone-integrated biosensors for pesticide detection is multifaceted, requiring a balanced focus on technical excellence, strategic intellectual property management, and rigorous regulatory compliance. The declining trend in patent applications after 2016 signals a market that is skeptical of promises and demands deliverable, robust, and user-centric solutions. Success depends on generating comprehensive validation data that addresses real-world variability, securing strong patents that protect core innovations, and engaging early with regulatory pathways. By adhering to these structured application notes and protocols, researchers and developers can significantly enhance the likelihood of translating a promising laboratory innovation into a trusted commercial product that ensures food safety and public health.
Market Analysis and Future Projections for Smart Biosensor Adoption
The global smart biosensor market is experiencing robust growth, fueled by technological advancements and increasing demand for real-time biological data across healthcare, environmental monitoring, and food safety sectors. Smart biosensors represent a transformative integration of biological recognition elements with transducers and digital technologies, enabling precise, real-time detection of specific biomarkers and analytes. These systems are increasingly leveraging smartphone integration for data processing, visualization, and communication, creating powerful decentralized diagnostic platforms [5]. The convergence of artificial intelligence (AI), Internet of Things (IoT) connectivity, and nanotechnology has accelerated the development of increasingly sophisticated biosensing solutions capable of addressing complex analytical challenges from point-of-care diagnostics to environmental pesticide detection [75].
The relevance of smart biosensors extends significantly into the field of visual pesticide detection, where they offer promising alternatives to traditional laboratory-based methods. Chromatography techniques coupled with mass spectroscopy, while highly sensitive and reliable, present several disadvantages including complex sample preparation protocols, high operational costs, time-consuming processes, and requirements for centralized laboratories and trained personnel, making them unsuitable for on-site pesticide detection [76]. Smart biosensors, particularly those integrated with mobile platforms, are emerging as viable solutions for rapid, sensitive, and quantitative monitoring of pesticide residues in food and water samples, providing the field-deployable tools urgently needed for environmental and food safety monitoring [76].
Global Market Analysis
Current Market Size and Growth Projections
The smart biosensor market demonstrates substantial growth potential across multiple related sectors. While specific market size data for smart biosensors alone is limited in the provided search results, related markets provide strong indicators of growth trajectories and commercial potential. The broader smart pest monitoring management system market, which incorporates biosensing technologies, was valued at USD 905.50 million in 2024 and is predicted to reach approximately USD 1,631.18 million by 2034, expanding at a compound annual growth rate (CAGR) of 6.07% from 2025 to 2034 [77].
Complementing this data, the pesticide detection market—a key application area for biosensors—is projected to grow from approximately USD 1.50 billion in 2025 to about USD 2.43 billion by 2035, reflecting a CAGR of 4.9% over the forecast period [78]. This growth is paralleled in specific technology segments like PPG biosensors, which are expected to grow at a remarkable 16.8% CAGR from 2025 to 2035, increasing from USD 648.5 million to USD 3,064.8 million [79]. These growth patterns collectively indicate strong market expansion for sensing technologies, with particularly rapid adoption expected in healthcare and environmental monitoring applications.
Table 1: Smart Biosensor-Related Market Size and Projections
| Market Segment | Base Year Value | Projection Year | Projected Value | CAGR |
|---|---|---|---|---|
| Smart Pest Monitoring Management System [77] | USD 905.50 million (2024) | 2034 | USD 1,631.18 million | 6.07% (2025-2034) |
| Pesticide Detection Market [78] | USD 1.50 billion (2025) | 2035 | USD 2.43 billion | 4.9% (2025-2035) |
| PPG Biosensors Market [79] | USD 648.5 million (2025) | 2035 | USD 3,064.8 million | 16.8% (2025-2035) |
Regional Adoption Trends
Geographic analysis reveals distinct regional patterns in technology adoption and market growth. North America currently dominates the smart pest monitoring management system market with the largest market share of 35% in 2024, supported by strong technological infrastructure, regulatory frameworks, and high adoption rates of IoT technologies [77]. The United States specifically leads in PPG biosensor adoption, driven by a robust ecosystem of wearable tech companies, high uptake of remote monitoring for chronic disease management, and established FDA-clearance pathways for digital diagnostics [79].
The Asia-Pacific region is expected to witness the fastest growth during the foreseeable period, fueled by rapid industrial expansion, increasing technology penetration, and growing awareness of health and environmental issues [77]. Countries like China, Japan, India, and South Korea are demonstrating particularly strong growth in biosensor adoption, with regional players providing end-to-end ecosystems combining cloud analytics, smart gadgets, and virtual consultations [79]. Europe maintains a steady market presence, with growth driven by strict regulations, sustainability goals, and strong industrial standards, particularly in Germany, the UK, France, Sweden, and the Netherlands [77] [79].
Table 2: Regional Market Analysis and Growth Trends
| Region | Market Position | Key Growth Drivers | Leading Countries/Areas |
|---|---|---|---|
| North America | Dominant market position (35% share in smart pest monitoring) [77] | Strong tech ecosystem, regulatory pathways, IoT adoption [77] [79] | United States, Canada [79] |
| Asia-Pacific | Fastest growing region [77] | Rapid industrialization, technology penetration, health awareness [77] [79] | China, Japan, India, South Korea [79] |
| Europe | Steady growth, established market [77] | Strict regulations, sustainability focus, digital health infrastructure [79] | Germany, UK, France, Netherlands [79] |
| Latin America/Middle East/Africa | Emerging markets [77] | Economic development, industrial expansion, infrastructure modernization [77] | Brazil, Mexico, UAE [77] |
Technology Segmentation and Adoption Patterns
The smart biosensor ecosystem encompasses diverse technological approaches, each with distinct adoption patterns. In the broader smart pest monitoring sector, hardware components held the dominant market share of 45% in 2024, as they form the essential foundation for data collection through smart traps and sensors [77]. However, the software segment is expected to witness the fastest growth, driven by AI-powered analytics that enable predictive pest identification, real-time monitoring, and optimized treatment timing [77].
In terms of detection technologies, IoT-based monitoring led the market with a significant share of 50% in 2024, attributable to its ability to provide real-time data, automate responses, increase efficiency, and promote sustainable pest control approaches [77]. The AI and vision systems segment is projected to grow at a notable CAGR, providing real-time, accurate pest identification and data analysis that leads to precise, data-driven management strategies [77]. For deployment models, cloud-based solutions held the largest market share of around 55% in 2024, offering scalability, cost-efficiency, real-time data analysis, and easy access via wireless platforms [77].
Key Market Drivers and Challenges
Growth Drivers and Opportunities
Several powerful forces are propelling the adoption of smart biosensor technologies. The rising global demand for sustainable and eco-friendly pest control methods represents a primary driver, fueled by regulations aimed at lowering pesticide usage and increasing focus on food safety and public health [77]. Growing organic farming practices and government initiatives promoting Integrated Pest Management (IPM) favor smart traps and sensors that enable precise pest control with minimal pesticide applications [77].
The integration of new technologies such as automated response systems presents substantial growth opportunities. The most promising future opportunity lies in integrating AI and IoT with automated response systems, enabling predictive, targeted, and sustainable management solutions [77]. Cloud computing platforms facilitate processing and analyzing large datasets, leading to more accurate, timely, and data-driven decisions across various sectors [77]. Advancements in detection technologies, including CRISPR/Cas12a-based platforms that have demonstrated limits of detection as low as 40 femtograms per reaction, are further expanding application possibilities [5].
The expanding scope of applications represents another significant growth driver. Smart biosensors now support diverse functions including healthcare monitoring (glucose, oxygen saturation, cardiac markers), public health (remote care, outbreak tracking), environmental safety (air/water quality assessment), food safety (contamination detection), and industrial biotech monitoring [5]. This application diversity ensures market expansion across multiple sectors rather than dependence on a single industry.
Adoption Challenges and Barriers
Despite promising growth, several challenges impede widespread biosensor adoption. Technical limitations present significant hurdles, particularly for smartphone-based biosensors which face issues with calibration inconsistencies, environmental variability, and limited interoperability across different smartphone models [5]. Sensor performance variability under real-world conditions further complicates deployment, as factors such as temperature fluctuations, humidity, and biological sample variability can distort readings, leading to diagnostic inaccuracies [5].
Economic factors also constrain market growth, including the high initial investment and ongoing maintenance costs associated with advanced biosensing technologies [77]. These cost barriers may deter potential users, especially in less affluent regions or smaller operations [77]. Additionally, the high cost of advanced equipment and the need for skilled personnel may constrain market growth in some regions [78].
Regulatory and integration challenges present further adoption barriers. The absence of unified communication standards and limited interoperability with electronic health records (EHRs) disrupts clinical workflows and impede seamless data exchange [5]. Regulatory uncertainty surrounding wellness versus diagnostic classification also precludes full clinical deployment in some markets [79]. Furthermore, at the production level, the high cost and limited scalability of advanced sensor components continue to restrict affordability and accessibility, particularly in low-resource settings [5].
Application Focus: Smartphone-Integrated Biosensors for Visual Pesticide Detection
Smartphone-integrated biosensors for pesticide detection represent a convergence of biological recognition elements, transducers, and mobile technology to create portable, sensitive detection platforms. These systems typically utilize enzymatic reactions, immunoassays, or nucleic acid-based detection mechanisms coupled with optical, electrochemical, or thermal transducers that convert biological interactions into quantifiable signals [5]. The smartphone serves as a power source, processor, display, and communication hub, enabling real-time, remote monitoring capabilities [5].
A prominent example of this technology is the integrated smartphone/resistive biosensor developed for organophosphate (OP) pesticide detection [76]. This biosensor leverages the hydrolytic activity of acetylcholinesterase (AChE) to its substrate, acetylcholine (ACh), and unique transport properties of polyaniline nanofibers (PAnNFs) within a chitosan/AChE/PAnNF/carbon nanotube (CNT) nanocomposite film on a gold interdigitated electrode [76]. The operating principle relies on OP pesticides inhibiting AChE, thus reducing the rate of ACh hydrolysis and consequently decreasing the rate of protons doping the PAnNFs. The resulting decrease in conductance of PAnNF is used to quantify OP pesticides in a sample [76].
Colorimetric detection approaches represent another significant methodology, utilizing digital image colorimetry (DIC) on smartphones to provide quantitative information about an analyte through color changes in a digital image [80]. These systems face challenges with variability between devices but can be standardized through appropriate calibration methodologies and controlled lighting conditions [80].
Diagram 1: Smartphone Biosensor Operational Workflow for Pesticide Detection
Experimental Protocol: Smartphone/Resistive Biosensor for OP Pesticide Detection
Objective: To detect and quantify organophosphate (OP) pesticides in food and environmental water samples using an integrated smartphone/resistive biosensor.
Materials and Equipment:
- Gold interdigitated electrodes (AuIDEs) with 20 μm features
- Acetylcholinesterase (AChE) enzyme
- Polyaniline nanofibers (PAnNFs) and carbon nanotubes (CNTs)
- Chitosan solution
- Acetylcholine (ACh) substrate
- Paraoxon-Methyl (PM) standard solutions
- Smartphone with custom-developed mobile application
- Portable digital multimeter for wireless data transmission
- Glass fiber (GF) pads (2.5 mm diameter)
- Parafilm template
- Vacuum desiccator
- Food/water samples (orange juice, grape juice, milk, meat, river water, well water)
Procedure:
Sensor Fabrication:
- Clean AuIDEs under light sonication in acetone followed by rinsing with ultrapure deionized water for 15 minutes each, then dry under vacuum.
- Attach a Parafilm template with a 2.5 mm diameter hole to the sensor surface to confine the sensing area.
- Treat the exposed sensing surface with 10 μL of 0.002% Tween-20 for 15 minutes, rinse with DI water, and dry in a vacuum desiccator overnight.
- Drop-cast 4 μL of partially-dedoped CNT/PAnNF suspension onto the exposed sensor surface and allow to air-dry.
- Place 2 μL of AChE solution on the CNT/PAnNF nanocomposite, allowing enzymes to enter the nanonetwork.
- Drop 2 μL chitosan onto the enzyme-entrapped nanocomposite and air-dry in a desiccator at 4°C.
- Peel off the parafilm template and store the prepared nanosensor in a desiccator at 4°C before use [76].
Preparation of Pre-loaded Pads:
- For signal generation pads: Cut GF membrane into 2.5 mm diameter circles, load with desired volume of ACh, then dry and store in a vacuum desiccator.
- For pre-treatment pads: Prepare similarly but load with anti-interference reagents (EDTA) and pH adjustment solution [76].
Sample Preparation:
- Prepare pesticide-spiked food and water samples according to established protocols [76].
- Adjust sample pH as needed using pre-treatment pads.
Measurement Procedure:
- Connect the biosensor to a portable digital multimeter for wireless data transmission to a smartphone.
- Apply sample to the biosensor platform along with the pre-treatment pad.
- Place the ACh-loaded signal generation pad in contact with the biosensor.
- Monitor conductance changes through the mobile application.
- Record measurements when signal stabilizes (typically within minutes).
- Analyze data using the smartphone application, which quantifies OP concentrations based on conductance reduction relative to inhibitor-free controls [76].
Data Analysis and Validation:
- The mobile application analyzes measurement data, displays testing results, and enables data sharing.
- Compare results with liquid chromatography-mass spectrometry validation methods.
- Calculate recovery rates from spiked samples; the developed biosensor demonstrated an average recovery rate of 98.3% with high reproducibility (RSD <5%) for Paraoxon-Methyl [76].
Table 3: Research Reagent Solutions for Smartphone Biosensor Pesticide Detection
| Reagent/Component | Function | Specifications |
|---|---|---|
| Acetylcholinesterase (AChE) [76] | Biorecognition element | Enzyme that hydrolyzes acetylcholine; inhibited by OP pesticides |
| Polyaniline Nanofibers (PAnNFs) [76] | Transducing element | Conducting polymer with tunable transport properties through doping/dedoping as function of pH |
| Carbon Nanotubes (CNTs) [76] | Signal amplifier | Enhance sensitivity and fast response times; part of nanocomposite film |
| Chitosan [76] | Enzyme immobilization matrix | Prevents enzyme leakage; biocompatible polymer |
| Acetylcholine (ACh) [76] | Enzyme substrate | Hydrolyzed by AChE to produce protons that dope PAnNFs |
| Gold Interdigitated Electrodes [76] | Sensor platform | Provide large transductive surface area; minimize gap length between finger electrodes |
| Glass Fiber Pads [76] | Reagent delivery | Pre-loaded with substrates/reagents for simplified testing |
Performance Metrics and Validation
The smartphone-integrated biosensor for OP pesticide detection demonstrates exceptional analytical performance. Under optimal conditions, the biosensor showed a wide linear range (1 ppt–100 ppb) with an exceptionally low detection limit (0.304 ppt) and high reproducibility (RSD <5%) for Paraoxon-Methyl as a model analyte [76]. When applied to spiked food and water samples, the biosensor provided an average recovery rate of 98.3% and delivered comparable results to liquid chromatography-mass spectrometry, confirming its reliability for real-sample analysis [76].
The platform addresses several critical challenges in biosensor development: sensitivity, stability, simplicity, portability, cost, and data sharing. The modification of AuIDE with CNT/PAnNF film acts as a signal amplifier, allowing for fast response times, improved stability, and high sensitivity for pesticide analysis [76]. The application of PAnNFs as the transducing element is essential due to its tunable transport properties through changes in doping/dedoping state as a function of pH [76]. The pre-dried reagent pads simplify the testing process, making it suitable for field applications.
Future Outlook and Strategic Recommendations
Emerging Trends and Technologies
The future of smart biosensor adoption will be shaped by several converging technological trends. Artificial intelligence and machine learning are increasingly being integrated into biosensing platforms, enabling enhanced diagnostic interpretation, predictive analytics, and personalized health insights [5]. Explainable AI is particularly important for clinical adoption, providing transparency in diagnostic decisions [5]. Advanced materials, particularly nanomaterials like gold nanoparticles and graphene, continue to improve signal transduction, with gold nanoparticles integrated into electrochemical biosensors shown to boost signal amplification efficiency by up to 50% [5].
Multimodal sensing approaches, combining optical, electrochemical, or thermal modalities, are improving diagnostic robustness through cross-validation and better accuracy [5]. Similarly, hybrid biosensor platforms that combine multiple sensing technologies (such as PPG with ECG or bioimpedance) are creating more comprehensive health monitoring solutions [79]. Innovations in power management, including self-powered systems using triboelectric generators or biochemical energy harvesters that operate without batteries, are expanding applications in low-resource or emergency contexts [5].
In the specific domain of pesticide detection, future developments will likely focus on multiplexed detection capabilities, allowing simultaneous screening for multiple pesticide classes in a single test. CRISPR-based detection systems, which have demonstrated limits of detection as low as 40 femtograms per reaction for specific DNA targets, may be adapted for pesticide detection through appropriate recognition elements [5]. Paper-based biosensors will continue to play important roles for affordable diagnostics, particularly in resource-limited settings [5].
Implementation Roadmap and Strategic Recommendations
For researchers and professionals working on smartphone-integrated biosensors for visual pesticide detection, several strategic recommendations emerge from the market analysis:
Focus on Integration and Interoperability: Develop biosensing platforms that seamlessly integrate with existing healthcare and environmental monitoring infrastructure, including electronic health records and public health surveillance systems. Address interoperability challenges through standardized communication protocols [5].
Prioritize User-Centered Design: Create solutions that address real-world usability challenges, including simplified calibration procedures, minimal sample preparation requirements, and intuitive user interfaces. This is particularly important for field applications in agricultural and environmental monitoring [5].
Advance Standardization and Validation: Establish standardized calibration protocols and performance validation frameworks to ensure reproducible and reliable results across different devices and platforms. This is essential for regulatory approval and clinical trust [5] [80].
Embrace Multidisciplinary Collaboration: Accelerate innovation through collaborations between material scientists, electrical engineers, software developers, and application domain experts (e.g., agricultural scientists, clinical chemists, environmental engineers).
Develop Sustainable Business Models: Consider tiered pricing models, subscription services for continuous monitoring, and partnerships with public health agencies to ensure economic viability and broad accessibility of biosensing technologies.
Diagram 2: Smart Biosensor Technology Adoption Roadmap
The market analysis and future projections presented indicate strong growth potential for smart biosensor technologies, particularly those integrated with mobile platforms for pesticide detection and other environmental monitoring applications. By addressing current technical challenges, focusing on user-centered design, and leveraging emerging technologies like AI and advanced nanomaterials, researchers and developers can capitalize on the significant market opportunities in this rapidly evolving field. The continued convergence of biological sensing, digital technology, and data science will likely yield increasingly sophisticated solutions that transform how we monitor and manage environmental health and safety.
Conclusion
Smartphone-integrated biosensors for visual pesticide detection represent a powerful convergence of biotechnology, nanomaterials, and digital innovation, poised to revolutionize on-site monitoring. The synthesis of foundational research, methodological advances, and rigorous validation underscores their potential to deliver rapid, sensitive, and cost-effective diagnostics. Key takeaways include the critical role of novel probes like UOFs and MIPs for selectivity, the necessity of AI-driven data analysis for accuracy across device platforms, and the importance of overcoming calibration and scalability challenges for widespread adoption. Future directions should focus on developing multi-analyte detection panels, creating fully self-powered systems, strengthening data security within IoT frameworks, and navigating regulatory pathways to transition these promising laboratory prototypes into reliable tools for ensuring global food safety, environmental health, and personalized medicine.
References
- 1. Research Progress of Biosensors in the Detection ... [https://www.mdpi.com/2079-6374/15/12/778]
- 2. Smartphone-Based Multiplexed Biosensing Tools for Health ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9406027/]
- 3. Smartphone-Based Biosensor Devices for Healthcare [https://pmc.ncbi.nlm.nih.gov/articles/PMC8945789/]
- 4. Bioreceptors for smartphone-based food contaminants ... [https://www.sciencedirect.com/science/article/abs/pii/S0166526X22001039]
- 5. Smartphone-Based Biosensors: Current Trends ... [https://www.mdpi.com/2673-4591/106/1/10]
- 6. Software | BAID-China - iGEM 2025 [https://2025.igem.wiki/baid-china/software]
- 7. Biosensors for Personal Mobile Health: A System ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7546526/]
- 8. Recent Advances in Smart Phone-Based Biosensors for ... [https://www.mdpi.com/2227-9040/13/7/221]
- 9. Smartphone biosensors - Nanosensors Group [https://nano.ece.illinois.edu/research-smartphone-biosensors/smartphone-biosensors/]
- 10. Development of integrated smartphone/resistive biosensor ... [https://www.sciencedirect.com/science/article/pii/S2590137023000997]
- 11. A fluorescence biosensor for organophosphorus pesticide ... [https://pubmed.ncbi.nlm.nih.gov/39486239/]
- 12. Best practices and current implementation of emerging ... [https://www.sciencedirect.com/science/article/pii/S0165993622003466]
- 13. ELISA Assay Technique | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-elisa.html]
- 14. ELISA: The Complete Guide [https://www.antibodies.com/applications/elisa]
- 15. Specific, Sensitive, and Quantitative Enzyme-Linked ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2730307/]
- 16. Aptamer-Based Fluorescent Biosensors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3205236/]
- 17. Aptamer-based high-sensitivity optical fiber SPR sensor for ... [https://www.sciencedirect.com/science/article/abs/pii/S092540052501038X]
- 18. Internet-of-things-integrated molecularly imprinted polymer ... [https://www.sciencedirect.com/science/article/abs/pii/S0269749124007437]
- 19. A comprehensive review on optical and electrochemical ... [https://www.frontiersin.org/journals/nutrition/articles/10.3389/fnut.2025.1596673/full]
- 20. Bioinspired HOF nanozyme-enhanced hydrogel biosensor ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400525015229]
- 21. Recent advances in nanomaterial-based optical ... [https://www.sciencedirect.com/science/article/pii/S2095177925001662]
- 22. Electrochemical Biosensors: A Prospective Insight to ... [https://www.sciencedirect.com/science/article/abs/pii/S2451910325001358]
- 23. An Integrated Nanosensor/Smartphone Platform for Point-of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12079637/]
- 24. Emerging trends in AI-integrated optical biosensors for point ... [https://pubs.rsc.org/en/content/articlehtml/2025/cc/d5cc04899k]
- 25. Advancing protein biosensors: redefining detection through ... [https://pubs.rsc.org/en/content/articlehtml/2025/ra/d4ra06791f]
- 26. Biosensors, Volume 15, Issue 9 (September 2025) [https://www.mdpi.com/2079-6374/15/9]
- 27. Emerging trends in AI-integrated optical biosensors for ... [https://pubmed.ncbi.nlm.nih.gov/41200923/]
- 28. Scientists Develop Smartphone Test to Detect Pesticides ... [https://www.spectroscopyonline.com/view/scientists-develop-smartphone-test-to-detect-pesticides-and-antibiotics-in-food]
- 29. Number of connected IoT devices growing 14% to 21.1 billion [https://iot-analytics.com/number-connected-iot-devices/]
- 30. Top 5 IoT Hardware Trends to Watch in 2025 [https://www.jaycon.com/top-5-iot-hardware-trends-to-watch-in-2025/]
- 31. Sensitivity, high throughput, and smart detection [https://www.sciencedirect.com/science/article/abs/pii/S0924424724001134]
- 32. Smartphone-Integrated Visual Inspection for Enhancing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11876467/]
- 33. Renewable uranium-based organic framework for removal ... [https://www.sciencedirect.com/science/article/abs/pii/S0022459625002257]
- 34. A smartphone-based fluorospectrophotometer and ratiometric ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10139973/]
- 35. Visual Strategies of Molecularly Imprinted Sensors for ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993625003401]
- 36. Ratiometric fluorescence sensor for organophosphorus ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566319307845]
- 37. A ratiometric fluorescent aptasensor for detection of ACE ... [https://www.sciencedirect.com/science/article/abs/pii/S1386142525012636]
- 38. A novel colorimetric strategy for rapid detection of ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400522003628]
- 39. Developing an in-house colorimetric method for detecting ... [https://chembioagro.springeropen.com/articles/10.1186/s40538-023-00442-3]
- 40. Fluorometric and colorimetric analysis of carbamate ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400519305684]
- 41. Fluorometric and Colorimetric Biosensors for the Assay of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12074314/]
- 42. Development of a Rapid and Sensitive Visual Pesticide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12346702/]
- 43. Smartphone-Integrated Electrochemical Devices for ... [https://www.mdpi.com/2079-6374/15/9/574]
- 44. Effective Soil Sampling for Sustainable Crop Production [https://extension.arizona.edu/publication/effective-soil-sampling-sustainable-crop-production]
- 45. Soil Testing Guide 2025 [https://alluvialsoillab.com/blogs/soil-analysis/soil-testing-guide-2025?srsltid=AfmBOoq0TcJyNd8USHie3tT182wyOl5F-9o8JQwaZQ1z-xBYzKAiSn7w]
- 46. Efficient Dual-Phase Visual Detection of Pesticides in Real ... [https://pubs.rsc.org/en/content/articlelanding/2025/ma/d5ma00630a]
- 47. Accelerated and precise identification of antioxidants ... [https://www.sciencedirect.com/science/article/abs/pii/S0039914024006544]
- 48. Development of machine learning enhanced low-cost ... [https://www.sciencedirect.com/science/article/abs/pii/S0263224125002490]
- 49. Recent developments in monitoring of organophosphorus ... [https://www.sciencedirect.com/science/article/pii/S2666154325000808]
- 50. A fluorescence biosensor for organophosphorus pesticide ... [https://www.sciencedirect.com/science/article/abs/pii/S1386142524014963]
- 51. Sensor Calibration at Ultralow Levels: Challenges and Solutions [https://www.environics.com/2025/07/07/sensor-calibration-at-ultralow-levels-challenges-and-solutions/]
- 52. which environmental stressors trigger calibration drift [https://www.clarity.io/blog/which-environmental-stressors-most-often-trigger-calibration-drift-and-necessitate-shorter-calibration-intervals]
- 53. To calibrate or not to calibrate, that is the question [https://www.sciencedirect.com/science/article/pii/S0043135422012830]
- 54. Multimodal Fusion-Driven Pesticide Residue Detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC12430378/]
- 55. Smartphone-Integrated Electrochemical Devices for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467198/]
- 56. Advanced visual sensing techniques for on-site detection ... [https://www.sciencedirect.com/science/article/pii/S2405844023011933]
- 57. Distance-Readout Visual Electrochemiluminescence ... [https://www.sciencedirect.com/science/article/abs/pii/S0925400525018544]
- 58. Development of a Rapid and Sensitive Visual Pesticide ... [https://www.mdpi.com/2304-8158/14/15/2628]
- 59. Protocol: a simple method for biosensor visualization of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9479286/]
- 60. Sciforum : Event management platform [https://sciforum.net/paper/view/22252]
- 61. Research Advances in Nanosensor for Pesticide Detection in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12298915/]
- 62. Efficient dual-phase visual detection of pesticides in real ... [https://pubs.rsc.org/en/content/articlehtml/2025/ma/d5ma00630a]
- 63. Bridging the Gap: The Role of Advanced Formulation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579833/]
- 64. An All-in-One Sustainable Smartphone Paper Biosensor for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12392259/]
- 65. Sensitivity and specificity [https://en.wikipedia.org/wiki/Sensitivity_and_specificity]
- 66. When lower isn't always better in biosensor research and ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566324006766]
- 67. Validation of an HPLC-MS/MS method for the quantification ... [https://www.sciencedirect.com/science/article/pii/S2590157524008265]
- 68. Determination of Pesticide Residues in Cucumber Using ... [https://www.waters.com/nextgen/us/en/library/application-notes/2022/determination-of-pesticide-residues-in-cucumber-using-gc-ms-ms-with-apgc-after-extraction-and-clean-up-using-quechers.html?srsltid=AfmBOoqxdi-Am-QeAESVJXSGFnBcJ4oSRoCNowJvy74c5bWiCguL4w9d]
- 69. Enzyme-Linked Immunosorbent Assay for the ... [https://www.sciencedirect.com/science/article/pii/S0362028X23063573]
- 70. Development of an ELISA for the detection of ... - PubMed - NIH [https://pubmed.ncbi.nlm.nih.gov/11368571/]
- 71. Comparison of Liquid Chromatography Mass Spectrometry ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7068296/]
- 72. Comparison of an HPLC-MS/MS Method with Multiple ... [https://openaccesspub.org/new-developments-in-chemistry/article/933]
- 73. Best practices and current implementation of emerging ... [https://www.sciencedirect.com/science/article/pii/S0165993623000730]
- 74. Digital Patient Monitoring Devices Patent Landscape ... [https://finance.yahoo.com/news/digital-patient-monitoring-devices-patent-164800236.html]
- 75. Smart Biosensor Market Industry Update 2025 [https://www.linkedin.com/pulse/smart-biosensor-market-industry-update-2025-prime-market-insight-iyqsf/]
- 76. Development of integrated smartphone/resistive biosensor for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10732357/]
- 77. Smart Pest Monitoring Management System Market Size to ... [https://www.precedenceresearch.com/smart-pest-monitoring-management-system-market]
- 78. Pesticide Detection Market Size & Trends 2025-2035 [https://www.futuremarketinsights.com/reports/pesticide-detection-market]
- 79. PPG Biosensors Market Size & Trends 2025 to 2035 [https://www.futuremarketinsights.com/reports/ppg-biosensors-market]
- 80. Solving Color Reproducibility between Digital Devices [https://www.mdpi.com/2079-6374/12/5/341]